Abstract
Background
Malaria is still a public health problem in Malaysia with chloroquine (CQ) being the first-line drug in the treatment policy of uncomplicated malaria. There is a scarcity in information about the magnitude of Plasmodium falciparum CQ resistance. This study aims to investigate the presence of single point mutations in the P. falciparum chloroquine-resistance transporter gene (pfcrt) at codons 76, 271, 326, 356 and 371 and in P. falciparum multi-drug resistance-1 gene (pfmdr1) at codons 86 and 1246, as molecular markers of CQ resistance.
Methods
A total of 75 P. falciparum blood samples were collected from different districts of Pahang state, Malaysia. Single nucleotide polymorphisms in pfcrt gene (codons 76, 271, 326, 356 and 371) and pfmdr1 gene (codons 86 and 1246) were analysed by using mutation-specific nested PCR and restriction fragment length polymorphism (PCR-RFLP) methods.
Results
Mutations of pfcrt K76T and pfcrt R371I were the most prevalent among pfcrt gene mutations reported by this study; 52% and 77%, respectively. Other codons of the pfcrt gene and the positions 86 and 1246 of the pfmdr1 gene were found mostly of wild type. Significant associations of pfcrt K76T, pfcrt N326S and pfcrt I356T mutations with parasitaemia were also reported.
Conclusion
The high existence of mutant pfcrt T76 may indicate the low susceptibility of P. falciparum isolates to CQ in Peninsular Malaysia. The findings of this study establish baseline data on the molecular markers of P. falciparum CQ resistance, which may help in the surveillance of drug resistance in Peninsular Malaysia.
Similar content being viewed by others
Background
Anti-malarial drug resistance is a major challenge to the control of falciparum malaria, the leading cause of morbidity and mortality especially in Africa and southern Asia[1]. The first P. falciparum chloroquine resistance was reported in the late 1950s in Southeast Asia along the Thai-Cambodian border[2, 3]. Further spread of CQ resistance was shown later to include neighboring countries in Asia, South America and Africa[4–8]. Moreover, P. falciparum has been also reported resistant to other anti-malarial drugs including sulphadoxine/pyrimethamine drug combination, mefloquine, atovaquone and artemisinin[9–12].
Three methods have been commonly used for monitoring anti-malarial drug efficacy and to screen for drug resistance; in vivo test, in vitro test and, more recently, the detection of molecular markers of anti-malarial drug resistance[13]. CQ resistance has been associated with point mutations in two genes; P. falciparum chloroquine resistance transporter (pfcrt) and multidrug resistance 1 (pfmdr1). It is well documented that the mutation at codon 76 of the pfcrt gene (pfcrt K76T), resulting in the substitution of threonine for lysine at position 76, is a key marker of P. falciparum CQ resistance[14]. However, the role of pfmdr1 point mutations on CQ resistance P. falciparum isolates remains a matter of debate.
In Malaysia, malaria is still a public health problem especially in the interior parts of the Peninsula and the states of Sabah and Sarawak (East Malaysia), with P. falciparum and Plasmodium vivax being the predominant species. A previous in vivo study followed by a molecular detection of resistance markers has been conducted in Sarawak and showed that CQ resistance occurred widely and therefore, CQ was replaced by sulphadoxine/pyrimethamine (SDX/PYR), and later by artemisinin combination therapy, as the drug of choice for treating uncomplicated falciparum malaria infections in East Malaysia[15]. In West Malaysia, CQ is still used as the first-line drug in the treatment policy of uncomplicated malaria caused by P. falciparum, although a previous in vivo study reported a CQ resistance rate of 63.6% among uncomplicated falciparum malaria patients[16]. Data on the molecular markers of CQ resistance in Peninsular Malaysia is not available. Therefore, this study was carried out to detect the molecular markers of anti-malarial drug resistance based on P. falciparum chloroquine resistance transporter (pfcrt) and P. falciparum multidrug resistance 1 (pfmdr1) genes.
Methods
Study area
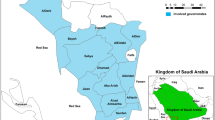
This study was carried out in Pahang state which is the largest state in Malaysia after Sabah and Sarawak (East Malaysia). The state is divided into 11 administrative districts and is located in the eastern North Kuala Lumpur with an area of 36,137 km2 and a total population of 1,443,365. The climate is equatorial with an average temperature of 21-32°C and an annual rainfall of between 2,000- 2,500 mm. Relative humidity in such areas is in the range of 80%. The economy is mostly agricultural; palm oil, natural rubber and timber are the major primary commodities. Malaria is endemic in Pahang, with P. falciparum and P. vivax accounting for the majority of reported cases.
Sampling and malaria microscopy
A total of 822 blood samples were collected from the different districts of Pahang (728 survey-based blood samples and 94 archived positive falciparum malaria slides) (Figure 1). The survey was conducted between May 2010 and May 2011 while the archived slides are for cases reported between 2007 and 2009. During the survey, finger prick blood samples were collected from participants in 20 villages of different districts of Pahang, and thick and thin blood films were prepared. Filter paper blood spots were also collected from each participant on 3MM Whatman® filter paper (Whatman International Ltd., Maidstone, England), and kept in clean, dry and well-sealed separate plastic bags for molecular analysis. Demographic data of the participants were collected by a standard semi structural questionnaire throughout face-to-face interview during the sampling survey. For archived slides, data was collected from the patients’ records.
Thick and thin blood films were stained with 10% diluted Giemsa and examined microscopically for the presence of malaria. About 200 fields under 1,000x magnification were examined from the thick film before the slide was considered negative. Parasitaemia was calculated by counting the asexual stages of the parasite against 300 white blood cells (WBC) and multiplied by 7500 as an assumed average total WBC count for the individuals[17]. Parasitaemia was categorized as low (< 1,000 parasites/μl of blood), moderate (1,000 – 9,999 parasites/μl of blood) and high (≥ 10,000 parasites/μl of blood). Archived malaria positive slides were also re-examined and parasitaemia were recorded.
Molecular identification of Plasmodium species
Genomic DNA was extracted from filter papers blood spots and from archived blood smears. Briefly, a disc of the filter paper blood spot (approximately 6 mm diameter) was punched out using a methanol-flamed puncher and placed in 1.5 ml centrifuge tubes using clean and flamed forceps. Archived slides were first cleaned with chloroform to remove oil and 50 ml of TE buffer was transferred onto the smear. The smear was completely wiped off the slide using small Whatman filter paper disc (cut by flamed-sterile puncher) and transferred into 1.5 ml microcentrifuge tubes. Genomic DNA was extracted using Qiagen blood and tissue kit (QIAGEN, DNeasy® Blood & Tissue Kit, Cat. no. 69506, Germany) according to the manufacurer’s instruction. DNA extract was finally eluted using 50 μL Qiagen AE elution buffer and kept at −20°C until used. Plasmodium species were identified using genus-specific and species-specific primers in nested PCR based on small subunit ribosomal RNA (18 s rRNA) gene[18]. PCR reaction and cycling conditions were conducted as mentioned previously[19].
Molecular detection of pfcrt and pfmdr1 mutations
The detection of mutations of different positions of pfcrt and pfmdr1 genes was performed using mutation-specific nested PCR and restriction fragment length polymorphism (PCR-RFLP) according to the protocols described previously[14, 20].
Primers for pfcrt K76T primary amplification were TCRP1 and TCRP2 while the secondary PCR was conducted by using the forward primer CRTP3 and the reverse primers TCRP4 m and TCRP4 w for mutant and wild types, respectively. Pfcrt Q271E alleles were amplified using primer set of CRT2a and CRT2b in the primary PCR and CRT271a and CRT271b in the secondary PCR. Amplification of N326S, I356T and R371I pfcrt mutations was carried out using the same primer set (CRT-3a and CRT-3b) in the first reaction, while secondary reactions were done using CRT326a and CRT326b, CRT356a and CRT356b, and CRT371a and CRT371b, respectively. A 20 μl reaction volume containing 8 μl of the secondary PCR amplicons and 1 unit of specific endonuclease digestion enzyme was incubated overnight at 37°C. The restriction enzymes XMN1, MSe1, AlwN1 and AflII were used for the digestion of pfcrt 271, 326, 356 and 371 amplicons while the mutant pfcrt allele T76 remained uncut.
For the detection of pfmdr1 gene mutations (N86Y and D1246Y), primers sets (MDR-A and MDR-B) and (1246-A and 1246-B) were used for the primary amplification of pfmdr1 86 and pfmdr1 1246, respectively while the secondary PCR was conducted using primers sets (MDR-D1 and MDR-D2) and (1246-D1 and 1246-D2) for flanking the mutation site of pfmdr1 86 and pfmdr1 1246 codons, respectively. Then, the Pfmdr1 N86Y and D1246Y amplicons were digested using Af1III and Bg1II restricted enzymes, respectively. The primers and PCR reagents were from iNtRON (iNtRON Biotechnology, Inc. Seoul, Korea). All PCR products were resolved by electrophoresis in a 2% agarose gel stained with SYBR safe and visualized by UV-transilluminator.
Genomic DNA of HB3, 3D7 and Dd2 strains of P. falciparum provided by Malaria Research and Reference Reagents Resources Centre (MR4, ATCC®, Manassas VA, USA) were used as positive controls for wild and mutant types, respectively.
Ethical consideration
The protocol of this study (Reference Number: 788.73) has been approved by the Medical Ethics Committee of the University of Malaya Medical Centre, Kuala Lumpur, Malaysia. The study was also registered with the National Medical Research Registry, Malaysia (Research ID: 5681). Before the commencement of sampling, the objectives of the study were explained to the people in their village setting and consents were obtained from those who have agreed to participate in the study.
Statistical analysis
Data was analysed using the SPSS statistical package version 13. Statistical associations between point mutations and parasitaemia were assessed using the Chi-square test with Yates' continuity correction. When the number of expected observations in one or more cells in a 2 × 2 contingency table is less than 5, Fisher’s Exact test was used. A p value ≤ 0.05 was considered statistically significant.
Results
Of the 822 collected samples, 728 were collected by sampling surveys in 20 villages of different districts of Pahang, and 94 were archived malaria positive slides from different health centres, hospitals and malaria control units in the same study area. Plasmodium falciparum parasites were detected in 75 of the total collected samples (64 from archived slides and 11 from surveys). Based on these 75 slides, the mean age of participants was 27.8 ± 4.1 years and 66% were males. The majority of participants were Malay (64.2%) followed by Aborigines (23.9), Indians (6.5%) and Chinese (2.7%). Moreover, 2 slides (2.7%) were from foreign workers who were residing in Malaysia for the past three years.
Plasmodium falciparum parasite density ranged from 50 asexual parasites/μl of blood to 17,100 with a parasitaemia geometric mean of 4,065 asexual parasites/μl. Moderate-to-high parasitaemic individuals (parasite count ≥ 1,000 parasites/μl of blood) were more frequent 58.7% while individuals with low parasitemia represented 41.3%.
All falciparum positive slides were examined for single nucleotide polymorphisms (SNP) at five positions of pfcrt gene (K76T, Q271E, N326S, I356T and R371I) and two positions of pfmdr1 genes (N86Y and D1246Y), and the results are shown in Figure 2. The detection rate was 100% for all positions of pfcrt and pfmdr1 markers except for 76 and 271 position of pfcrt (98.7%); one sample was negative for each of them. Of pfcrt point mutations, pfcrt K76T and pfcrt R371I had higher mutant alleles than other positions (52% and 77%, respectively). Other positions showed predominance for the wild type with rates of 93%, 88% and 76% for the pfcrt Q271, pfcrt N326 and pfcrt I356, respectively. Moreover, polymorphism analysis of pfmdr1 revealed a high predominance of wild type for both markers; 95% and 96% for pfmdr1 N86 and pfmdr1 D1246, respectively.
The association of mutations at different codons of pfcrt and pfmdr1 with the parasitaemia, age of participant, gender, race and year of infection was examined (Table 1). Of the five positions of mutation at pfcrt gene, mutations at codon 76, 326 and 356 showed significant associations with moderate-to- high parasitaemia as compared to low parasitaemia (p < 0.01). On the other hand, the association of pfcrt mutations with age, race, gender of participants and year of infection was not significant (p > 0.05). Similarly, the association of pfmdr1 gene mutations at positions 86 and 1246 with the tested variables was not significant.
Discussion
This is the first study in Peninsular Malaysia on the detection of molecular markers of pfcrt and pfmdr1 for chloroquine and other anti-malarial drugs resistance. The high prevalence rate (52%) of pfcrt K76T point mutation reported by the present study reflects low susceptibility of P. falciparum to the CQ. This is in agreement with previous studies in Malaysia and neighbouring countries including Thailand, Indonesia and Philippine[15, 16, 21–23]. In Sarawak (East Malaysia), Cox-Singh et al. reported a strong association of pfcrt K76T with in vitro CQ resistance; pfcrt K76T was found higher at a fixation level (100%) in all isolates resistant to CQ[15]. Similar detection rates of mutant pfcrt T76 with a significant association with in vitro CQ resistance had also been reported in Thailand and Philippine[21, 23].
Previous studies in other countries have showed such association between pfcrt K76T mutation and in vivo resistance of P. falciparum to CQ. In Mali, the presence of pfcrt K76T mutation was reported in all infections occurred within two weeks of administration of chloroquine while it was absent in all isolates that responded well to CQ[14, 24]. Similar findings were reported in Uganda, Sudan and Yemen[19, 25, 26]. In the same vein, a previous study aimed at comparing two areas with different susceptibility levels to CQ found that the prevalence of wild type of pfcrt K76 was higher in areas with lower CQ resistance, while the mutant type was higher in areas with higher CQ resistance[27].
Except for pfcrt 371, the findings showed that other examined codons of the pfcrt gene (271, 326 and 356) were found mostly of wild type represented 93%, 88% and 76%, respectively. Moreover, pfcrt R371I mutation was detected in 77% of the isolates. Previous reports had suggested a correlation between CQ resistance and the accumulation of point mutations in the pfcrt gene[28, 29]. However, the role of these mutations in conferring CQ resistance is unclear and suggested to have a complementary effect in maintaining the functional property of transporter protein[14, 30]. Thus, further studies to explain the role of these mutations including pfcrt R371I are required.
Regarding pfmdr1, the present study showed a predominance of wild type of both pfmdr1 N86 and pfmdr1 D1246 (95% and 96%, respectively). The role of pfmdr1 gene mutations in anti-malarial drugs resistance is still controversial. Previous in vivo studies reported the absence of mutations at codons pfmdr1 86 and 1246 in the CQ resistant infections[31–33]. Similarly, a previous study in south-eastern Iran reported a strong association between pfcrt K76T, but not pfmdr1 N86Y mutation and in vivo CQ resistance[34]. In contrast, a recent study from Madagascar reported an association between pfmdr1 Y86 mutant alleles and CQ clinical resistance with no such association with pfcrt gene[35]. Moreover, an in vitro study showed that pfmdr1 mutations in P. falciparum can confer resistance to high levels of chloroquine, and that these pfmdr1 mutations has an important role in the resistance of P. falciparum to mefloquine and quinine[36].
The high prevalence of wild alleles of pfmdr1 N86 and pfmdr1 D1246 could be considered as an indicator for low susceptibility of P. falciparum isolates to mefloquine, amodiaquine and quinine[21, 37–40]. On the other hand, previous studies have found increased sensitivity to the anti-malarials mefloquine and artemisinin in P. falciparum isolates with mutations in the pfmdr1 gene[41, 42]. However, a previous in vivo study in Thailand found that treatment failure with mefloquine is significantly associated with increased pfmdr1 copy number but not with the single point mutations[43].
The present study found a significant association between moderate-to-high parasitaemia and pfcrt point mutations at codons 76, 326 and 356. This is in agreement with previous studies in Tanzania, Sudan, Nigeria and Yemen[19, 44, 45]. However, studies from Sudan and Gabon showed no significant association between the pfcrt point mutations and severity of malaria[46, 47]. Moreover, a recent study in the Yemen found higher frequency of pfcrt K76T mutation in P. falciparum isolates from patients aged > 10 years as compared to younger individuals[19]. On the other hand, a previous study among Nigerian children aged ≤ 12 years with acute uncomplicated P. falciparum malaria showed that the association between the presence of both mutant pfcrt T76 and pfmdr1 Y86 alleles with in vivo anti-malarial drug resistance was age dependent[38]. However, the present study found no significant association between gene point mutations and age of patients. Most of the positive samples in this study were from adult patients aged 22–40 years, and this limited range of age may constraint examining such association.
Conclusions
The high prevalence of mutation at codon 76 of the pfcrt gene (pfcrt K76T) is a key indicator of P. falciparum CQ resistance spread in Peninsular Malaysia. A further study to examine the implemented malaria drug policy using PCR-corrected anti-malarial drug efficacy trail in large sample set nationally is recommended. Continuous molecular surveillance using pfcrt and pfmdr1 genes as molecular markers of chloroquine resistance are highly recommended in both East and West Malaysia, for monitoring the efficacy of chloroquine in Peninsular Malaysia and for the re-emergence of wild-type of P. falciparum in East Malaysia (Sarawak and Sabah).
References
WHO: World Malaria Report 2010. 2010, Geneva: World Health Organization
Young MD, Contacos PG, Stitcher JE, Millar JW: Drug resistance in Plasmodium falciparum from Thailand. Am J Trop Med Hyg. 1963, 12: 305-314.
Harinasuta T, Suntharasamai P, Viravan C: Chloroquine resistant falciparum malaria in Thailand. Lancet. 1965, ii: 657-660.
Grimmond TR, Donovan KO, Riley ID: Chloroquine resistant malaria in Papua New Guinea. P N G Med J. 1976, 19: 184-185.
Moore DV, Lanier JE: Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. 1961, 10: 5-9.
Fogh S, Jepsen S, Effersoe P: Chloroquine-resistant Plasmodium falciparum malaria in Kenya. Trans R Soc Trop Med Hyg. 1979, 73: 228-229. 10.1016/0035-9203(79)90220-7.
Onori E: The problem of Plasmodium falciparum drug resistance in Africa south of the Sahara. Bull World Health Organ. 1984, 62 (Suppl): 55-62.
Overbosch D, van den Wall Bake AW, Stuiver PC, van der Kaay HJ: Chloroquine-resistant falciparum malaria from Malawi. Trop Geogr Med. 1984, 36: 71-72.
Peters W: Chemotherapy and drug resistance in malaria, 2nd ed edn. 1987, New York: Academic Press, Inc.
Nosten F, Kuile F, Chongsuphajaisiddhi T, Luxemburger C, Webster HK, Edstein M, Phaipun L, Thew KL, White NJ: Mefloquine-resistant falciparum malaria on the Thai-Burmese border. Lancet. 1991, 337: 1140-1143. 10.1016/0140-6736(91)92798-7.
Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ: Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am J Trop Med Hyg. 1996, 54: 62-66.
Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ: Artemisinin resistance in Plasmodium falciparum malaria. N Eng J Med. 2009, 361: 455-467. 10.1056/NEJMoa0808859.
Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR: Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002, 2: 209-218. 10.1016/S1473-3099(02)00239-6.
Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV: A molecular marker for chloroquine-resistant falciparum malaria. N Eng J Med. 2001, 344: 257-263. 10.1056/NEJM200101253440403.
Cox-Singh J, Lu HY, Davis TME, Ilett KF, Hackett LP, Matusop A, Singh B: Application of a multi-faceted approach for the assessment of treatment response in falciparum malaria: A study from Malaysian Borneo. Int J Parasitol. 2003, 33: 1545-1552. 10.1016/S0020-7519(03)00199-1.
Hakim SL, Roohi SWAS, Zurkurnai Y, Rain AN, Mansor SM, Palmer K, Navaratnam V, Mak JW: Plasmodium falciparum: increased proportion of severe resistance (RII and RIII) to chloroquine and high rate of resistance to sulfadoxine-pyrimethamine in Peninsular Malaysia after two decades. Trans R Soc Trop Med Hyg. 1996, 90: 294-297. 10.1016/S0035-9203(96)90258-8.
Dicko A, Sagara I, Djimde AA, Toure SO, Traore M, Dama S, Diallo AI, Barry A, Dicko M, Coulibaly OM, Rogier C, de Sousa A, Doumbo OK: Molecular markers of resistance to sulphadoxine-pyrimethamine one year after implementation of intermittent preventive treatment of malaria in infants in Mali. Malar J. 2010, 9: 9-10.1186/1475-2875-9-9.
Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah M, Rahman H: A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999, 60: 687-692.
Al-Mekhlafi AM, Mahdy MA, Al-Mekhlafi HM, Azazy AA, Fong MY: High frequency of Plasmodium falciparum chloroquine resistance marker (pfcrt T76 mutation) in Yemen: an urgent need to re-examine malaria drug policy. Parasit Vectors. 2011, 4: 94-10.1186/1756-3305-4-94.
Lopes D, Rungsihirunrat K, Nogueira F, Seugorn A, Gil JP, do Rosario VE, Cravo P: Molecular characterisation of drug-resistant Plasmodium falciparum from Thailand. Malar J. 2002, 1: 12-10.1186/1475-2875-1-12.
Rungsihirunrat K, Chaijareonkul W, Seugorn A, Na-Bangchang K, Thaithong S: Association between chloroquine resistance phenotypes and point mutations in pfcrt and pfmdr1 in Plasmodium falciparum isolates from Thailand. Acta Trop. 2009, 109: 37-40. 10.1016/j.actatropica.2008.09.011.
Maguire JD, Susanti AI, Krisin Sismadi P, Fryauff DJ, Baird JK: The T76 mutation in the pfcrt gene of Plasmodium falciparum and clinical chloroquine resistance phenotypes in Papua, Indonesia. Ann Trop Med Parasitol. 2001, 95: 559-572. 10.1080/00034980120092516.
Hatabu T, Iwagami M, Kawazu S, Taguchi N, Escueta AD, Villacorte EA, Rivera PT, Kano S: Association of molecular markers in Plasmodium falciparum crt and mdr1 with in vitro chloroquine resistance: A Philippine study. Parasitol Int. 2009, 58: 166-170. 10.1016/j.parint.2009.01.010.
Djimde A, Doumbo O, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Cortese J, Su X, Fidock D, Nomura T, Wellems T, Plowe C: In vivo chloroquine responses and pfcrt polymorphisms in Bandiagara, Mali. Am J Trop Med Hyg. 1999, 61 (supplement): 335-336.
Dorsey G, Kamya MR, Singh A, Rosenthal PJ: Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis. 2001, 183: 1417-1420. 10.1086/319865.
Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D: High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J Infect Dis. 2001, 183: 1535-1538. 10.1086/320195.
Kyosiimire-Lugemwa J, Nalunkuma-Kazibwe AJ, Mujuzi G, Mulindwa H, Talisuna A, Egwang TG: The Lys-76-Thr mutation in pfcrt and chloroquine resistance in Plasmodium falciparum isolates from Uganda. Trans R Soc Trop Med Hyg. 2002, 96: 91-95. 10.1016/S0035-9203(02)90252-X.
Durrand V, Berry A, Sem R, Glaziou P, Beaudou J, Fandeur T: Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) and their relationship to chloroquine resistance in vitro. Mol Biochem Parasitol. 2004, 136: 273-285. 10.1016/j.molbiopara.2004.03.016.
Lim P, Chy S, Ariey F, Incardona S, Chim P, Sem R, Denis MB, Hewitt S, Hoyer S, Socheat D, Merecreau-Puijalon O, Fandeur T: Pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob Agents Chemother. 2003, 47: 87-94. 10.1128/AAC.47.1.87-94.2003.
Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LMB, Sidhu AS, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE: Mutations in the Plasmodium falciparum digestive vacuole transmembrane protein pfcrt and evidence for their role in chloroquine resistance. Mol Cell. 2000, 6: 861-871. 10.1016/S1097-2765(05)00077-8.
Awad-El-Kariem FM, Miles MA, Warhurst DC: Chloroquine-resistant Plasmodium falciparum isolates from the Sudan lack two mutations in the pfmdr1 gene thought to be associated with chloroquine resistance. Trans R Soc Trop Med Hyg. 1992, 86: 587-589. 10.1016/0035-9203(92)90140-8.
Basco LK, Bras JL, Rhoades Z, Wilson CM: Analysis of pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from Subsaharan Africa. Mol Biochem Parasitol. 1995, 74: 157-166. 10.1016/0166-6851(95)02492-1.
von Seidlein L, Duraisingh MT, Drakeley CJ, Bailey R, Greenwood BM, Pinder M: Polymorphism of the pfmdr1 gene and chloroquine resistance in Plasmodium falciparum in the Gambia. Trans R Soc Trop Med Hyg. 1997, 91: 450-453. 10.1016/S0035-9203(97)90281-9.
Zakeri S, Afsharpad M, Kazemzadeh T, Mehdizadeh K, Shabani A, Djadid ND: Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2008, 78: 633-640.
Andriantsoanirina V, Ratsimbasoa A, Bouchier C, Tichit M, Jahevitra M, Rabearimanana S, Raherinjafy R, Mercereau-Puijalon O, Durand R, Menard D: Chloroquine clinical failures in Plasmodium falciparum malaria are associated with mutant pfmdr-1, not pfcrt in Madagascar. PLoS One. 2010, 5: e13281-10.1371/journal.pone.0013281.
Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF: Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000, 403: 906-909. 10.1038/35002615.
Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, White NJ, Nosten F, Krishna S: The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999, 43: 2943-2949.
Happi CT, Gbotosho GO, Folarin OA, Bolaji OM, Sowunmi A, Kyle DE, Milhous W, Wirth DF, Oduola AM: Association between mutations in Plasmodium falciparum chloroquine resistance transporter and Plasmodium falciparum multidrug resistance 1 genes and in vivo amodiaquine resistance in Plasmodium falciparum malaria-infected children in Nigeria. Am J Trop Med Hyg. 2006, 75: 155-161.
Tinto H, Guekoun L, Zongo I, Guiguemde RT, D’Alessandro U, Ouedraogo JB: Chloroquine-resistance molecular markers (pfcrt T76 and pfmdr-1 Y86) and amodiaquine resistance in Burkina Faso. Trop Med Int Health. 2008, 13: 238-240. 10.1111/j.1365-3156.2007.01995.x.
Baliraine FN, Nsobya SL, Achan J, Tibenderana JK, Talisuna AO, Greenhouse B, Rosenthal PJ: Limited ability of Plasmodium falciparum pfcrt, pfmdr1, and pfnhe1 polymorphisms to predict quinine in vitro sensitivity or clinical effectiveness in Uganda. Antimicrob Agents Chemother. 2011, 55: 615-622. 10.1128/AAC.00954-10.
Duraisingh MT, Roper C, Walliker D, Warhurst DC: Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol. 2000, 36: 955-961. 10.1046/j.1365-2958.2000.01914.x.
Sidhu ABS, Valderramos SG, Fidock DA: Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005, 57: 913-926. 10.1111/j.1365-2958.2005.04729.x.
Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S: Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004, 364: 438-447. 10.1016/S0140-6736(04)16767-6.
Khalil IF, Alifrangis M, Tarimo DS, Staalso T, Satti GMH, Theander TG, Ronn AM, Bygbjerg IC: The roles of the pfcrt 76 T and pfmdr1 86Y mutations, immunity and the initial level of parasitaemia, in predicting the outcome of chloroquine treatment in two areas with different transmission intensities. Ann Trop Med Parasitol. 2005, 99: 441-448. 10.1179/136485905X46441.
May J, Meyer CG: Association of Plasmodium falciparum chloroquine resistance transporter variant t76 with age-related plasma chloroquine levels. Am J Trop Med Hyg. 2003, 68: 143-146.
Giha HA, Elbashir MI, A-Elbasit IE, A-Elgadir TME, ElGhazali GE, Mackinnon MJ, Babiker HA: Drug resistance-virulence relationship in Plasmodium falciparum causing severe malaria in an area of seasonal and unstable transmission. Acta Trop. 2006, 97: 181-187. 10.1016/j.actatropica.2005.10.004.
Mayengue P, Kalmbach Y, Issifou S, Kremsner P, Ntoumi F: No variation in the prevalence of point mutations in the pfcrt and pfmdr1 genes in isolates from Gabonese patients with uncomplicated or severe Plasmodium falciparum malaria. Parasitol Res. 2007, 100: 487-493.
Acknowledgements
The authors wish to acknowledge the fruitful cooperation of the Directors of Health Offices in the districts of Pahang, Mr. Mohd Mahir from the Vector Control Unit-Kuala Lipis, Mr. Shamsuddin bin Mohd Nasib from Pahang State Health Department, and Mr. Muhammad Azhan bin Abdul Majid and Mr. Hood bin Adam from Raub Health Office. We thank MR4 (Malaria research and Reference Reagents Resources Centre) for providing us with genomic DNA of Plasmodium falciparum positive controls (3D7, HB3 and dD2) contributed by (MR4, ATCC®, Manassas, VA, USA). This work was financially supported by University of Malaya Research Grant (RG151/09HTM) and University of Malaya High Impact Research Fund, HIR-MOHE (H-20001-00-E00051).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WMA was involved in all phases of the study, including study design, data collection, data analysis, interpretation, and write-up of the manuscript; HMA, MAKM and JS designed the study; WMA and HMA performed the statistical analysis; MAKM provided technical advisory support in genotyping and data interpretation; All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Atroosh, W.M., Al-Mekhlafi, H.M., Mahdy, M.A. et al. The detection of pfcrt and pfmdr1 point mutations as molecular markers of chloroquine drug resistance, Pahang, Malaysia. Malar J 11, 251 (2012). https://doi.org/10.1186/1475-2875-11-251
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-11-251