Abstract
Background
We retrieved different reports containing different genetic effects of − 1082 A/G, − 819 T/C, and − 592 A/C polymorphisms within the IL-10 (interleukin-10) gene on the susceptibility to clinical atopic dermatitis.
Methods
Herein, we conducted a meta-analysis to comprehensively assess such a genetic relationship after collecting the available published evidence. STATA 12.0 software was used for the statistical analysis under the allelic, homozygotic, heterozygotic, dominant, recessive and carrier genetic models.
Results
By retrieving and screening database literature, a total of 16 eligible case-control studies were finally selected. For the IL-10 -1082 A/G polymorphism, we did not detect a significant difference between atopic dermatitis cases and population-based controls in the overall meta-analysis under the genetic models of allele G vs. A (P = 0.540), GG vs. AA (P = 0.853), AG vs AA (P = 0.265), AG + GG vs AA (P = 0.221), GG vs AA+AG (P = 0.540) and carrier G vs. A (P = 0.643). Moreover, a statistically non-significant association was observed in the most subgroup meta-analyses by the factors of ethnicity, country and Hardy-Weinberg equilibrium. Likewise, the negative results were detected for the synthetic analysis of IL-10 -819 T/C and − 592 C/A polymorphisms.
Conclusion
The current evidence does not support a strong genetic relationship between IL-10 -1082 A/G, − 819 T/C and − 592 A/C polymorphisms and the susceptibility to atopic dermatitis.
Similar content being viewed by others
Background
Clinical atopic dermatitis is a common and chronic inflammatory disorder of the epithelial barrier with relapsing eczematous lesions and intense irritation [1,2,3]. The pathogenesis of atopic dermatitis remains unclear. The skin microbiome, genetics and innate/adaptive immune responses are closely related to the incidence and development of atopic dermatitis [4,5,6,7,8,9]. For example, a genome-wide association study (GWAS) of 246 recalcitrant atopic dermatitis patients and 551 negative controls from the Korean population reported susceptibility loci within the 13q21.31 region [9]. In the current study, we are interested in quantitatively investigating the possible effect of interleukin-10 (IL-10) polymorphisms on susceptibility to atopic dermatitis.
The IL-10 cytokine participates in the modulation of acquired immune and anti-inflammatory responses [10, 11]. Within the promoter region of the IL-10 gene, three common single nucleotide polymorphisms (SNPs), namely, − 1082 A/G (rs1800896), − 819 T/C (rs1800871) and − 592 A/C (rs1800872), were identified [12,13,14]. Different results regarding the genetic effect of IL-10 polymorphisms on the risk of atopic dermatitis were reported by individual researchers. For example, the IL-10 -1082 A/G polymorphism was reportedly linked to susceptibility to atopic dermatitis in patients from India [15], Italy [16], and the Czech Republic [17]. Nevertheless, the IL-10 -1082 A/G polymorphism was not associated with atopic dermatitis risk in the Korean population [18] or Polish population [19]. In addition, the TT genotype frequency of the IL-10 -819 T/C polymorphism in atopic dermatitis patients was higher than that in healthy control subjects in India [15]. However, the CC genotype of the IL-10 -819 T/C polymorphism was also reported to be associated with an enhanced risk of severe atopic dermatitis in patients from the Czech Republic [17]. A negative association between the IL-10 -819 T/C polymorphism and atopic dermatitis risk was also reported in Saudi Arabia [20] and Taiwan of China [21]. This issue merits the performance of a relative meta-analysis.
Some meta-analyses examined the variable degrees of the effect of IL-10 SNPs on several skin-associated clinical diseases. For instance, the IL-10 -819 T/C polymorphism, rather than − 1082 A/G and − 592 A/C, was reported to be associated with a decreased risk of skin cancer [22]. In 2013, Chen, et al. conducted a meta-analysis to assess the possible impact of the IL-10 -1082 A/G polymorphism on the risk of atopic dermatitis [23]. Along with the enrolment of new publications, it is important to conduct an updated meta-analysis to reassess the role of the IL-10 polymorphisms in the risk of atopic dermatitis.
Methods
Database retrieval
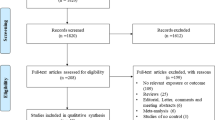
Our meta-analysis follows the guide of preferred reporting items for systematic reviews and meta-analyses (PRISMA) [24]. The PRISMA-based flow chart is shown in Fig. 1. In April 2019, five online electronic databases, including PubMed, Embase, Web of Science (WOS), China National Knowledge Infrastructure (CNKI) and WANFANG, were used to retrieve data by combining the different terms of “IL-10”, “atopic dermatitis” and “polymorphism”. The search strategy for PubMed containing the Medical Subject Headings (MeSH) and entry terms was as follows: ((((((((Interleukin-10 [MeSH Terms]) OR Interleukin 10) OR IL10) OR IL-10) OR CSIF-10) OR Cytokine Synthesis Inhibitory Factor)) AND (((((((((((((((((Dermatitis, Atopic [MeSH Terms]) OR Atopic Dermatitides) OR Atopic Dermatitis) OR Dermatitides, Atopic) OR Neurodermatitis, Atopic) OR Atopic Neurodermatitides) OR Atopic Neurodermatitis) OR Neurodermatitides, Atopic) OR Neurodermatitis, Disseminated) OR Disseminated Neurodermatitides) OR Disseminated Neurodermatitis) OR Neurodermatitides, Disseminated) OR Eczema, Atopic) OR Atopic Eczema) OR Eczema, Infantile) OR Infantile Eczema))) AND ((((((Polymorphism, Genetic [MeSH Terms]) OR Polymorphisms, Genetic) OR Genetic Polymorphisms) OR Genetic Polymorphism) OR Polymorphism (Genetics)) OR Polymorphisms (Genetics)). The search strategy for Embase containing the Emtree and Synonyms: (‘atopic dermatitis’/exp. OR ‘atopic eczema’ OR ‘coca sulzberger disease’ OR ‘coca sulzberger syndrome’ OR ‘dermatitis, atopic’ OR ‘eczema atopica’ OR ‘eczema endogenous’ OR ‘eczema infantum’ OR ‘eczema, infantile’ OR ‘endogenous eczema’ OR ‘infantile eczema’ OR ‘neurodermatitis constitutionalis’ OR ‘neurodermatitis disseminata’ OR ‘neurodermatitis, atopic constitutional’) AND (‘il10’ OR ‘csif’ OR ‘cytokine synthesis inhibitory factor’ OR ‘il 10’ OR ‘il-10’ OR ‘interleukin-10’) AND (‘genetic polymorphism’/exp. OR ‘polymorphism (genetics)’ OR ‘polymorphism, genetic’).
Study selection
After database retrieval, we designed our inclusion and exclusion criteria to select the eligible case-control studies for synthetic analysis. Exclusion criteria: (a) other diseases; (b) other genes; (c) animal/cell assay; (d) meeting abstract; (e) meta-analysis; (f) review paper; (g) other IL-10 variant; and (h) duplicate study. Inclusion criteria: (a) case and control study; (b) atopic dermatitis; (c) -1082 A/G, − 819 T/C, − 592 A/C polymorphisms within the IL-10 gene; and (d) the allelic or genotypic frequency data of cases and controls.
Data extraction
Subsequently, we extracted basic information, including first author, publication time (year), ethnicity, SNPs, genotypic or allelic frequency data in both controls and cases, control source, genotyping method, sample size, and P value of Hardy-Weinberg equilibrium (HWE). The Newcastle-Ottawa quality assessment Scale (NOS) scores of study quality were also measured.
Statistical analysis
After referring to several reported meta-analyses [25,26,27], we utilized STATA 12.0 software (Stata Corporation, College Station, TX, USA) to conduct the I2 test and Q statistic test (for heterogeneity evaluation), DerSimonian-Laird and Mantel-Haenszel method (for association test), Begg’s test and Egger’s test (for publication bias evaluation) [28, 29], and sensitivity analysis (for the assessment of data stability). The high heterogeneity level was considered to perform the DerSimonian-Laird method under a random-effect model when I2 was larger than 50% or the P value was less than 0.05. In contrast, the Mantel-Haenszel method under a fixed-effect model was used for the association test. In addition, the data of the odds ratio (OR), 95% confidence interval (CI) and P value were calculated under the allelic, homozygotic, heterozygotic, dominant, recessive and carrier genetic models in the overall meta-analysis as well as the relative subgroup analysis by the factors of ethnicity, country and HWE. Only the pooling results from at least three case-control studies were considered in our study.
Results
Enrolled studies
As shown in Fig. 1, we initially identified a total of 222 studies, including PubMed (n = 28), Embase (n = 79), WOS (n = 104), CNKI (n = 1) and WANFANG (n = 10), and we removed the 82 duplicates. Additionally, we ruled out another 116 studies by the exclusion criteria shown in Fig. 1. Moreover, after the eligibility assessment, we eliminated another six articles without the genotypic or allelic frequency data in cases or controls, and two articles regarding the other IL-10 variant were excluded. As a result, a total of 16 eligible case-control studies [15,16,17,18,19,20,21, 30,31,32,33,34,35,36,37,38] with the summarized basic information in Table 1 were enrolled. We observed population-based controls and high-quality controls (Additional file 1: Table S1, all NOS scores greater than five) in each study.
Meta-analysis of IL-10 -1082 a/G SNP
During the meta-analysis of the IL-10 -1082 A/G polymorphism (Table 2), 14 case-control studies were enrolled for the models of allele G vs. A (1,593 cases/2095 controls) and AG + GG vs. AA (1536 cases/2041 controls); 13 studies (1499 cases/1909 control) for the AG vs AA and carrier G vs. A; 11 studies (1288 cases/1742 controls) for the models of GG vs. AA and GG vs. AA+AG. In the association test shown in Table 2, we did not detect a significant difference between atopic dermatitis cases and population-based controls in the overall meta-analysis under the genetic models of allele G vs. A (P = 0.540), GG vs. AA (P = 0.853), AG vs AA (P = 0.265), AG + GG vs AA (P = 0.221), GG vs AA+AG (P = 0.540) and carrier G vs. A (P = 0.643). Likewise, we observed a statistically non-significant association in the subgroup analysis of “Asian”, “Caucasian”, “Poland”, and “Y, with a P value of HWE >0.05” (Table 2, P > 0.05), only apart from the “Asian” subgroup under the GG vs AA+AG model (P = 0.003, OR = 2.22). However, when we excluded the studies with the P value of HWE < 0.05, the negative results were obtained in the subgroup analysis of “Asian/Y”, “Caucasian/Y” and “Poland/Y” (Table 2, all P > 0.05). Forest plot data of overall meta-analysis under the allelic model (Fig. 2a) and subgroup analysis by ethnicity under allelic (Additional file 2: Figure S1), heterozygotic (Additional file 2: Figure S2) and dominant (Additional file 2: Figure S3) models were provided. In summary, the IL-10 -1082 A/G SNP may have no genetic effect on the risk of atopic dermatitis.
Meta-analysis of IL-10 -819 T/C SNP
In addition, for the IL-10 -819 T/C SNP (Table 3), nine case-control studies (1228/1544) were included in the allelic model, while eight studies (1134/1358) were included in other models. As shown in Table 2, negative results were detected in cases compared with that in population-based controls during the overall meta-analysis under the models of allele C vs. T (P = 0.547), CC vs. TT (P = 0.588), TC vs. TT (P = 0.405), TC + CC vs. TT (P = 0.433), CC vs. TT + TC (P = 0.821), and carrier C vs. T (P = 0.801). Likewise, we observed a non-significant association in the subgroup analysis of “Asian”, “Y” and “Asian/Y”, under all genetic models (Table 3, all P > 0.05). The relative forest plots are displayed in Fig. 3a (overall meta-analysis under the allelic model), Additional file 2: Figure S4 (subgroup analysis by ethnicity under the allelic model), Additional file 2: Figure S5 (subgroup analysis by ethnicity under the heterozygotic model), and Additional file 2: Figure S6 (subgroup analysis by ethnicity under the dominant model). Our findings suggested that the IL-10 -819 T/C SNP may not be associated with the risk of atopic dermatitis.
Meta-analysis of IL-10 -592 a/C SNP
For the IL-10 -592 A/C SNP (Table 4), nine case-control studies (1032/1363) were enrolled in the allelic model, while eight studies (938/1177) were in enrolled in other models. As shown in Table 4, we only observed positive results in the overall meta-analysis under AC vs. AA (P = 0.039, OR = 0.77) and AC + CC vs. AA (P = 0.026, OR = 0.76) as well as in the subgroup analysis of “Asian” under allele C vs. A (P = 0.033, OR = 0.85) and AC + CC vs. AA (P = 0.049, OR = 0.77) but not others (all P > 0.05). It is worth mentioning that the negative results were observed in the “Asian/Y” subgroup, under all genetic models, when only the studies with the P value of HWE > 0.05 were included in the analysis of “Asian” subgroup. The relative forest plots are displayed in Fig. 4a and Additional file 2: Figures S7-S9. The above outcomes did not provide evidence of the strong association between IL-10 -592 A/C SNP and atopic dermatitis susceptibility.
Heterogeneity, publication bias and sensitivity analysis
As shown in Additional file 1: Table S2, a random-effect model (DerSimonian and Laird method) was used under the heterozygotic and dominant models in the meta-analysis of the IL-10 -1082 A/G SNP; allelic, homozygotic, heterozygotic, dominant and recessive models of the − 819 T/C SNP; and allelic, homozygotic, recessive models of the − 592 A/C SNP (all I2 > 50% or P < 0.05). In addition, we did not observe a large publication bias in most above analyses (Additional file 1: Table S3, P > 0.05), only apart from the Egger’s test under the allelic (P = 0.028) and heterozygotic (P = 0.021) models of the − 819 T/C SNP. Egger’s publication bias plots are displayed in Figs. 2b, 3b and 4b. Our sensitivity analyses in Figs. 2c, 3c and 4c also suggested the statistical stability of pooling outcomes under the allelic models. Similar data were observed in other models (data not shown).
Discussion
In 2013, Chen, et al. included seven case-control studies [17,18,19, 21, 32, 34, 35] to perform a meta-analysis of the association between the IL-10 -1082 A/G polymorphism and the risk of atopic dermatitis [23]. In the present study, we collected the available data from diverse sources and finally added another eight new case-control studies [15, 16, 20, 30, 31, 33, 36, 37] to conduct another updated meta-analysis under the genetic models of allele A vs. G, GG vs. AA, AG vs AA, AG + GG vs AA, GG vs AA+AG and carrier A vs. G. Subgroup analyses stratified by ethnicity, country and HWE were also conducted. Our data failed to support the strong genetic link between the IL-10 -1082 A/G SNP and the risk of atopic dermatitis, which is in line with the outcomes of the above meta-analyses. Recently, another meta-analysis by Zhao, et al. [39] reported the same result on the potential association between IL-10 -1082 A/G SNP and atopic dermatitis susceptibility under the recessive model in the Association population. However, only three case-control studies [20, 30, 37] were enrolled in this comparison, and P value of HWE < 0.05 in controls of two studies [20, 37] were detected.
With regards to IL-10 -819 T/C SNP, a meta-analysis containing nine case-control studies was performed under the six genetic models. The subgroup analysis by the ethnicity was also performed, when the studies with the P value of HWE < 0.05 were removed. We found that IL-10 -819 T/C SNP may not be strongly linked to the risk of atopic dermatitis. Even though the potential positive conclusion was observed under the recessive model in the Caucasian populations [39], only two case-control studies [17, 35] were enrolled for synthetic analysis. The linkage disequilibrium between the IL-10 -819 T/C and − 592 C/A polymorphisms exists in some reports [15, 21]. However, different genotype frequencies were also reported in other reports [17, 18, 35, 37]. Here, we also pooled the available data to perform a meta-analysis, which did not strongly support the genetic relationship between the IL-10 -592 C/A SNP and the risk of atopic dermatitis. This is also in line with the conclusion of Zhao, et al. [39].
Our meta-analysis exhibits several advantages. First, all eligible studies contain population-based negative control subjects. Second, our results of Begg’s and Egger’s tests ruled out the presence of large publication bias. Third, our sensitivity analyses support the stability of pooling results.
Despite this, some disadvantages still may limit our statistical evaluation. First, as in other meta-analyses, a small sample size was enrolled in some analyses. For example, less than ten case-control studies were included in the overall meta-analysis of the IL-10 -819 T/C and − 592 C/A polymorphisms and all the stratification analyses. For the IL-10 -819 T/C SNP, only two case-control studies [17, 35] were included in the subgroup analysis of “Caucasian”. Only three case-control studies were obtained in the subgroup analysis of “Poland” for IL-10 -1082 A/G polymorphism, and less than three studies were for the other subgroup analyses of “Country”, such as “China” or “India”. Second, a high heterogeneity level among studies was observed in some overall meta-analyses. Third, the genotypic distribution of control groups in some studies was not in line with the Hardy-Weinberg equilibrium. Fourth, of the eligible case-control studies, only allelic frequency data were extracted in one study [21], and the combined genotypic frequency (AG + GG) of the IL-10 -1082 A/G polymorphism was obtained in another study [31]. Fifth, even though we obtained haplotypic data for the − 1082 A/G, − 819 T/C, − 592 A/C polymorphisms within the IL-10 genes, including “ACC”, “ATA”, and “GCC”, very limited data resulted in the failure of related meta-analysis. Sixth, one meta-analysis reported that the − 590 C/T polymorphism within the IL-4 (interleukin-4) gene may be linked to the risk of atopic dermatitis, especially for Asian children [40]. Considering the extensive role of interleukins (ILs) in the immune responses of diverse organisms [41,42,43,44,45], it is important to analyse the combined effects of different interleukin gene variants in either the resistance or susceptibility to genetic disorders, including many skin disorders, for a more objective and comprehensive assessment. However, we focused on the role of three SNPs of the IL-10 gene, one member in the interleukin gene family, in the susceptibility to atopic dermatitis in this study, based on the limitations of study publications. Given the contextual effects of the polymorphism and the diversity of the heterozygotic response, more interleukin gene variant-associated association investigations by additional adjusted clinical or environmental factors should be conducted when we can obtain access to more association investigation data for years to come.
Conclusion
Overall, based on the current case-control association data, IL-10 -1082 A/G, − 819 T/C and − 592 A/C polymorphisms may not be strongly related to atopic dermatitis susceptibility, which would be greatly strengthened by a larger sample size.
Abbreviations
- ARMS:
-
amplification refractory mutational system
- ASPCR:
-
allele-specific PCR
- CI:
-
confidence interval
- CNKI:
-
China National Knowledge Infrastructure
- FLG:
-
filaggrin
- GWAS:
-
genome-wide association study
- HWE:
-
Hardy-Weinberg equilibrium
- IL-10:
-
Interleukin-10
- KASP:
-
Kompetitive Allele Specific PCR
- NOS:
-
Newcastle-Ottawa quality assessment Scale
- OR:
-
odds ratio
- PB:
-
population-based control
- PCR:
-
polymerase chain reaction
- PRISMA:
-
systematic reviews and meta-analyses
- RFLP:
-
restriction fragment-length polymorphism
- SSP:
-
sequence specific primer
- WOS:
-
Web of Science
References
Tollefson MM, Bruckner AL. Atopic dermatitis: skin-directed management. Pediatrics. 2014;134(6):e1735–44.
Tsakok T, Woolf R, Smith CH, Weidinger S, Flohr C. Atopic dermatitis: the skin barrier and beyond. Br J Dermatol. 2018: Jul 3. https://doi.org/10.1111/bjd.16934 [Epub ahead of print].
Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers. 2018;4(1):1.
Sala-Cunill A, Lazaro M, Herraez L, Quinones MD, Moro-Moro M, Sanchez I. Skin allergy Committee of Spanish Society of a, clinical I. basic skin care and topical therapies for atopic dermatitis: essential approaches and beyond. J Investig Allergol Clin Immunol. 2018;28(6):379–91.
Tamari M, Hirota T. Genome-wide association studies of atopic dermatitis. J Dermatol. 2014;41(3):213–20.
Powers CE, McShane DB, Gilligan PH, Burkhart CN, Morrell DS. Microbiome and pediatric atopic dermatitis. J Dermatol. 2015;42(12):1137–42.
Bin L, Leung DY. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin Immunol. 2016;12:52.
Stemmler S, Hoffjan S. Trying to understand the genetics of atopic dermatitis. Mol Cell Probes. 2016;30(6):374–85.
Kim KW, Myers RA, Lee JH, Igartua C, Lee KE, Kim YH, Kim EJ, Yoon D, Lee JS, Hirota T, et al. Genome-wide association study of recalcitrant atopic dermatitis in Korean children. J Allergy Clin Immunol. 2015;136(3):678–84 e4.
Lobo-Silva D, Carriche GM, Castro AG, Roque S, Saraiva M. Balancing the immune response in the brain. IL-10 and its regulation. 2016;13(1):297.
Breen EC. Pro- and anti-inflammatory cytokines in human immunodeficiency virus infection and acquired immunodeficiency syndrome. Pharmacol Ther. 2002;95(3):295–304.
Shin HD, Park BL, Kim LH, Kim JS, Kim JW. Interleukin-10 haplotype associated with total serum IgE in atopic dermatitis patients. Allergy. 2005;60(9):1146–51.
Qian BX, Ye Q, Zhao XY, Han T, Wang FM, Yang J. Meta-analysis of the relation between IL10 promoter polymorphisms and autoimmune liver disease risk. Genet Test Mol Biomarkers. 2018;22(5):302–13.
Harsini S, Saghazadeh A, Nedjat S, Rezaei N. Associations between interleukin-10 polymorphisms and susceptibility to juvenile idiopathic arthritis: a systematic review and meta-analysis. Eur Cytokine Netw. 2018;29(1):16–26.
Jain C, Das S, Ramachandran VG, Saha R, Bhattacharya SN, Dar S. Malassezia yeast and cytokine gene polymorphism in atopic dermatitis. J Clin Diagn Res. 2017;11(3):DC01–DC5.
Esposito S, Patria MF, Spena S, Codeca C, Tagliabue C, Zampiero A, Lelii M, Montinaro V, Pelucchi C, Principi N. Impact of genetic polymorphisms on paediatric atopic dermatitis. Int J Immunopathol Pharmacol. 2015;28(3):286–95.
Kayserova J, Sismova K, Zentsova-Jaresova I, Katina S, Vernerova E, Polouckova A, Capkova S, Malinova V, Striz I, Sediva A. A prospective study in children with a severe form of atopic dermatitis: clinical outcome in relation to cytokine gene polymorphisms. J Investig Allergol Clin Immunol. 2012;22(2):92–101.
Sohn MH, Song JS, Kim KW, Kim ES, Kim KE, Lee JM. Association of interleukin-10 gene promoter polymorphism in children with atopic dermatitis. J Pediatr. 2007;150(1):106–8.
Lesiak A, Kuna P, Zakrzewski M, van Geel M, Bladergroen RS, Przybylowska K, Stelmach I, Majak P, Hawro T, Sysa-Jedrzejowska A, et al. Combined occurrence of filaggrin mutations and IL-10 or IL-13 polymorphisms predisposes to atopic dermatitis. Exp Dermatol. 2011;20(6):491–5.
Bin Huraib G, Al Harthi F, Arfin M, Al-Sugheyr M, Rizvi S, Al-Asmari A. Cytokine gene polymorphisms in Saudi patients with atopic dermatitis: a case-control study. Biomark Insights. 2018;13:1177271918777760.
Chang YT, Lee WR, Yu CW, Liu HN, Lin MW, Huang CH, Chen CC, Lee DD, Wang WJ, Hu CH, et al. No association of cytokine gene polymorphisms in Chinese patients with atopic dermatitis. Clin Exp Dermatol. 2006;31(3):419–23.
Zhao H, Yang J, Yu Z, Shen H, Huang X, Zhang M, Long T, Cailing A, Wang W. Synthetic analysis of associations between IL-10 polymorphisms and skin cancer risk. Oncotarget. 2018;9(6):6728–36.
Chen S, Zhao L. Association between the IL10 -1082A>G (rs1800896) single-nucleotide polymorphism and atopic dermatitis: a systematic review and meta-analysis. Dermatitis. 2013;24(4):161–5.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Zhang YQ, Ding W, Xu L, Zhang L, Han Z, Jiang Q, Wang Z, Jin S. Meta-analysis of association between TCF7L2 polymorphism rs7903146 and type 2 diabetes mellitus. BMC Med Genet. 2018;19(1):38.
Yao J, Qi XL, Zhang Y. The Alu-insertion progesterone receptor gene polymorphism is not associated with breast cancer: a meta-analysis. BMC Med Genet. 2018;19(1):16.
Qu Y, Li S, Zhao F, Wang Y, Mu D, Ma WQ. Association of genetic polymorphisms in vascular endothelial growth factor with susceptibility to coronary artery disease: a meta-analysis. BMC Med Genet. 2018;19(1):108.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–34.
Yinji X, Jinping T, Huijuan W. Association of IL-10 gene polymorphism with atopic dermatitis. Basic and Clinical Medicine. 2010;30(7):763–6.
Babic Z, Sabolic Pipinic I, Varnai VM, Kezic S, Macan J. Associations of TNFalpha -308G>a, TNFalpha -238G>a, IL-1alpha -889C>T and IL-10 -1082G>a genetic polymorphisms with atopic diseases: asthma, rhinitis and dermatitis. Int Arch Allergy Immunol. 2016;169(4):231–40.
Arkwright PD, Chase JM, Babbage S, Pravica V, David TJ, Hutchinson IV. Atopic dermatitis is associated with a low-producer transforming growth factor beta(1) cytokine genotype. J Allergy Clin Immunol. 2001;108(2):281–4.
Lesiak A, Zakrzewski M, Przybylowska K, Rogowski-Tylman M, Wozniacka A, Narbutt J. Atopic dermatitis patients carrying G allele in −1082 G/a IL-10 polymorphism are predisposed to higher serum concentration of IL-10. Arch Med Sci. 2014;10(6):1239–43.
Reich K, Westphal G, Konig IR, Mossner R, Schupp P, Gutgesell C, Hallier E, Ziegler A, Neumann C. Cytokine gene polymorphisms in atopic dermatitis. Br J Dermatol. 2003;148(6):1237–41.
Stavric K, Peova S, Trajkov D, Spiroski M. Gene polymorphisms of 22 cytokines in Macedonian children with atopic dermatitis. Iran J Allergy Asthma Immunol. 2012;11(1):37–50.
Zakrzewski M, Lesiak A, Przybyłowska K, Stelmach I, Kuna P, Sysa-Jȩdrzejowska A, Narbutt J. The lack of association between −590 C/T IL-4 and -1082 a/G IL-10 gene polymorphisms and the development of atopic dermatitis. Przeglad Dermatologiczny. 2010;97(4):253–9.
Behniafard N, Amirzargar AA, Gharagozlou M, Delavari F, Hosseinverdi S, Sotoudeh S, Farhadi E, Mahmoudi M, Khaledi M, Moghaddam ZG, et al. Single nucleotide polymorphisms of the genes encoding IL-10 and TGF-β1 in Iranian children with atopic dermatitis. Allergol Immunopathol. 2018;46(2):155–9.
Sheng W, Yang L, Kang K. IL10 gene polymorphisms in patients with atopic dermatitis of Han population in Tangshan. Shangdong Med J. 2018;58(32):29–32.
Zhao J, Chen ZY, Li LF. Association between the IL-10-1082G/a, IL-10-592A/C, and IL-10-819G/a polymorphisms and atopic dermatitis susceptibility: a meta-analysis. Genet Test Mol Biomarkers. 2019. https://doi.org/10.1089/gtmb.2018.0276 Epub ahead of print.
Liang J, Liu Y, Xue R, Chen L, Chen H, Shao L, Wang J, Zhang X. Interleukin 4 -590C/T (rs2243250) polymorphism is associated with increased risk of atopic dermatitis: meta-analysis of case-control studies. Dermatitis. 2017;28(2):144–51.
Moorlag S, Roring RJ, Joosten LAB, Netea MG. The role of the interleukin-1 family in trained. Immunity. 2018;281(1):28–39.
Kawamura T, Ogawa Y, Aoki R, Shimada S. Innate and intrinsic antiviral immunity in skin. J Dermatol Sci. 2014;75(3):159–66.
Burmeister AR, Marriott I. The Interleukin-10 family of cytokines and their role in the CNS. Front Cell Neurosci. 2018;12:458.
Su LC, Liu XY, Huang AF, Xu WD. Emerging role of IL-35 in inflammatory autoimmune diseases. Autoimmun Rev. 2018;17(7):665–73.
Kaplanski G. Interleukin-18: biological properties and role in disease pathogenesis. Immunol Rev. 2018;281(1):138–53.
Acknowledgements
The authors gratefully thanked the editors and reviewers to help improve the manuscript, and the American Journal Experts (https://www.aje.cn/) for professional help with English usage in this manuscript.
Funding
The study design, database searching and data interpretation were supported by a grant from National Nature Science Foundation of China (31170830).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Author information
Authors and Affiliations
Contributions
YQ and JH designed the study. YQ, JK and JH extracted, analyzed, and interpreted the data. YQ and JH drafted the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declared that they had no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. NOS assessment system. Table S2. Heterogeneity evaluation. Table S3. Publication bias assessment. (DOCX 43 kb)
Additional file 2:
Figure S1. Subgroup analysis of the IL-10 -1082 A/G polymorphism according to ethnicity under the allele G vs. A model. Figure S2. Subgroup analysis of the IL-10 -1082 A/G polymorphism according to ethnicity under the AG vs. AA model. Figure S3. Subgroup analysis of the IL-10 -1082 A/G polymorphism according to ethnicity under the AG + GG vs. AA model. Figure S4. Subgroup analysis of the IL-10 -819 T/C polymorphism according to ethnicity under the allele C vs. T model. Figure S5. Subgroup analysis of the IL-10 -819 T/C polymorphism according to ethnicity under the TC vs. TT model. Figure S6. Subgroup analysis of the IL-10 -819 T/C polymorphism according to ethnicity under the TC + CC vs. TT model. Figure S7. Subgroup analysis of IL-10 -592 A/C polymorphism according to ethnicity under the allele C vs. A model. Figure S8. Subgroup analysis of IL-10 -592 A/C polymorphism according to ethnicity under the AC vs. AA model. Figure S9. Subgroup analysis of IL-10 -592 A/C polymorphism according to ethnicity under the AC + CC vs. AA model. (ZIP 2797 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Qi, Y., Kong, J. & He, J. Genetic relationship between IL-10 gene polymorphisms and the risk of clinical atopic dermatitis. BMC Med Genet 20, 83 (2019). https://doi.org/10.1186/s12881-019-0817-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-019-0817-8