Abstract
Background
The role of progesterone receptor (PGR) gene polymorphisms in breast cancer is still controversial. Here, we performed a meta-analysis to determine whether the Alu insertion is associated with an increased risk of breast cancer and, further, whether the Alu insertion contributes to the development of breast cancer.
Methods
Using database searches, we selected 10 controlled case studies that met a rigorous set of inclusion criteria; these studies included 2106 cases and 1660 controls. We generated odds ratios and 95% confidence intervals in order to determine the strength of the relationship between the Alu insertion and breast cancer incidence. We also performed additional subgroup analyses and sensitivity analyses to further clarify the relationship.
Results
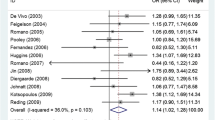
Using a random effects model, we concluded that the Alu insertion was not associated with the risk of breast cancer under the dominant genetic model; the pooled OR was 1.025 (95% CI = 0.526–1.994, p = 0.943). When a subgroup analysis was performed according to ethnicity, we found that the Alu insertion was associated with breast cancer incidence in Indians and Indo-European mixed racial groups, but the association disappeared for patients of Caucasian or Latino decent.
Conclusions
Our meta-analysis showed that the Alu-insertion progesterone receptor gene polymorphism was not associated with breast cancer. These results provide further information regarding the association between the Alu insertion in the PGR gene and the incidence of breast cancer.
Similar content being viewed by others
Background
Breast cancer is the leading cause of cancer-related deaths in women worldwide; globally, approximately 1 million women experience breast cancer each year [1, 2]. Current research demonstrates that an interaction between multiple abnormal genetic and environmental factors can generate a susceptibility to breast cancer [3]. A multitude of factors have been postulated to influence the development of breast cancer including age, ethnicity, early or delayed menarche, use of oral contraceptives, breastfeeding, and age at menopause as well as a number of genetic factors [4, 5]. Nevertheless, a definite etiology of breast cancer has not yet been identified.
The human progesterone receptor, encoded by the PGR gene, is a member of the steroid receptor superfamily. The progesterone receptor is essential for mediating the effects of progesterone, which is critical for the establishment and maintenance of pregnancy. Intracellular PGRs and their associated protein kinase C molecules are known to regulate tumor cell proliferation and metastasis, such as during the infiltration process of human glioblastomas [6]. Moreover, membrane PGR have been shown to mediate most non-classical progesterone actions; as such, membrane PGRs may be useful as pharmacologic targets or biomarkers of cancer and other reproductive diseases [7]. In addition, increased expression of PGRs has been reported in gastric cancer [8]. Signature patterns of expression of the estrogen and PGR signaling pathways may be used to predict prognosis and guide management of colorectal cancer [9]. Therefore, polymorphisms in PGR that effect its level of expression may be associated with variations in the risk of breast cancer. Several reports have attempted to address the relationship between PGR expression and breast cancer risk in populations of various ethnicities, although with mixed results [10,11,12,13,14].
The Alu element, a short interspersed nuclear element, is the most successful retrotransposon in primate genomes; it exists at an estimated copy number of 1.1 million in the human genome [15]. Each Alu element is approximately 300 bp in length and has a dimeric structure. Because of their abundance and sequence identity, Alu elements are frequently involved in genomic rearrangements within the human genome. Genomic rearrangements can result in genetic disorders such as Alport syndrome, Fabry disease, and peeling skin disease [16,17,18,19]. In fact, Alu elements are related to about 0.1% of human genetic disorders [20]. Recently a young human-specific 306 bp Alu insertion was found in the PGR gene, in intron G between exons 7 and 8. The Alu insertion contains a half-ERE/SPI site, which may disproportionately increase the transcription of PGR after estrogen stimulus [21]. Some studies reported that this particular Alu insertion was associated with breast cancer [22, 23], while others did not observe the association [24,25,26].
These conflicting results are likely due to the limited sample size found in the studies as well as differing genetic backgrounds. Meta-analysis is widely-used in medicine as a statistical method of reconciling studies with inconsistent results [27]. Therefore, we carried out a meta-analysis of studies investigating the relationship between the Alu insertion and the risk of breast cancer.
Methods
Selection and inclusion criteria for relevant studies
We searched three online electronic databases (Embase, PubMed, and Web of Science) in order to identify potential studies for inclusion in our meta-analysis; the data of the last search update was November 2016. We used the following key words in our literature searches: progesterone receptor, PROGINS, Alu insertion, breast cancer, and mammary. Articles cited by the potential studies and relevant review articles were also checked for additional supplementary studies. The following inclusion criteria were utilized: (1) a case-control study design; (2) patients with a diagnosis of breast cancer; and (3) inclusion of the allele and/or genotype frequencies. In cases where studies included the same or overlapping data, we chose the most recent article. The corresponding authors were contacted for other useful information (such as sample characteristics) and additional data not presented in the initial article.
Data extraction
Two researchers (Jun Yao and Xing-ling Qi) independently collected the relevant data, from all eligible publications. If there was a disagreement, a consensus was obtained after discussion. The following characteristics were collected from each study chosen for inclusion: first author’s last name, publication year, region, ethnicity, numbers of each genotype of both cases and controls, and source of controls.
Statistical analysis
Our meta-analysis was completed using Stata version 10.0 (Stata Corp., College Station, TX). We used a chi-square test to determine the Hardy-Weinberg equilibrium (HWE) of each genotype frequency in control subjects. Thakkinstian’s method was used to calculate pooled frequency analyses [28]. All statistical tests were two-sided and p < 0.05 was considered statistically significant.
In order to measure the strength of the association between breast cancer and the Alu insertion, we calculated odd’s ratios (ORs) with 95% confidence intervals (Cis). We used a random effects model to pool the effect sizes across studies. This model measure the possible effect size across populations with different genetic backgrounds after considering the heterogeneity among the included studies [29]. An allele contrast model, a dominant model, and a recessive model were used to calculate overall pooled ORs. Using A as the risk allele, we compared OR1 (AA vs. aa), OR2 (Aa vs. aa), and OR3 (AA vs. Aa); these pairwise differences were used to select the most appropriate genetic model under the instruction as previously described [27, 30].
We also determined the degree of heterogeneity across studies using the Q-statistic; p > 0.05 indicated a lack of heterogeneity and p < 0.05 indicated the presence of heterogeneity [31, 32]. I2 was the proportion of detected variation in effect size due to the actual discrepancies across studies; classically, the I2 statistic is used to define low (< 25%), moderate (~ 50%), and high (> 75%) levels of heterogeneity [33]. We also performed subgroup analyses by ethnicity (i.e. Caucasian, African-American, Latino, Indian, and Indo-European) as well as by source of control subjects (i.e. hospital-based vs. population-based).
We used a funnel plot to estimate publication bias. The standard error of log(OR) for each study was plotted vs. its log(OR); asymmetric plots indicated potential publication bias. The degree of asymmetry was measured using Egger’s test; p < 0.05 was considered significant publication bias [34].
Finally, we performed sensitivity analyses to measure the potential influence of each study on the final pooled effect size as previously described [35].
Results
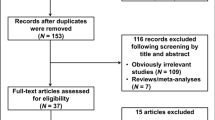
After excluding published works with overlapping data and those that did not meet inclusion criteria, the final meta-analysis included a total of nine published articles containing 10 studies including 2106 cases and 1660 controls [22,23,24,25,26, 36,37,38,39] (Fig. 1). Key characteristics of the included studies are described in Table 1. The frequencies of each genotype and allele along with their HWE values are listed in Table 2. Of the 10 studies, only one study showed significant deviation from the expected HWE (p = 0.0390) [23].
We calculated the pooled frequencies of the Alu insertion in the control populations stratified by ethnicity. T1/T1 represents homozygote individuals lacking the 306 bp Alu insertion, T1/T2 represents heterozygotes with the insertion, and T2/T2 indicates homozygous individuals with the Alu insertion. The allelic frequency of the Alu insertion varied across ethnicities: the pooled T2 allele frequency was highest among Indians (17.7%, 95% CI = 17.3–18.2%), followed by Caucasians (14.6%, 95% CI = 10.5–18.8%), Latinos (14.1%, 95% CI = 14.0–14.1%), African-Americans (4.9%, 95% CI = 4.9–5.0%), and Indo-Europeans (1.4%, 95% CI = 1.4–1.4%). The overall pooled T2 allele frequency was 12.6% (95% CI = 7.4–17.8%).
The association between the Alu insertion and the risk of breast cancer was determined in all 2106 cases and 1660 control subjects from 10 studies using pooled ORs and the corresponding 95% CIs for the homozygous codominant, heterozygous codominant, dominant, recessive, and allele contrast genetic models (Table 3). Finally, we selected the dominant model according to the principle of genetic model selection [30, 40]. The results indicated that there was no association between the Alu insertion and the occurrence of breast cancer (Fig. 2). For the dominant model, we used the random effects model to calculate a pooled OR of 1.025 (95% CI = 0.526–1.994, p = 0.943). Ethnicity subgroup analysis indicated that the Alu insertion was associated with breast cancer in Indian (OR = 0.091, CI = 0.033–0.254, p < 0.001) and Indo-European patients (OR = 11.620, 95% CI = 5.331–25.327, p < 0.001), but no association was found in Caucasians (OR = 0.916, 95% CI = 0.673–1.243, p = 0.576) or Latinos (OR = 1.712, 95% CI = 0.898–3.263, p = 0.102) (Table 4). Furthermore, we found no association between the Alu insertion and the risk of breast cancer in a subgroup analysis by source of controls (population-based: OR = 1.179, 95% CI = 0.515–2.699, p = 0.697; hospital-based: OR = 0.635, 95% CI = 0.385–1.046, p = 0.075).
Sensitivity analysis
Sensitivity analysis was conducted to measure the influence of each study on the resulting meta-analysis. The relevant pooled ORs indicated that no significant change appeared when each study was omitted, one at a time, from the overall meta-analysis. Thus, the final pooled results are both stable and reliable.
Publication bias
The potential publication bias was evaluated using a funnel plot (Fig. 3). An Egger’s test was also utilized as a supplementary test of bias; results of this test also indicated no publication bias (p = 0.889).
Discussion
Our meta-analysis included 10 studies, comprised of 2106 cases and 1660 controls, that investigated the association between PGR gene polymorphism (specifically, the Alu insertion) and the occurrence of breast cancer. Overall, the results of our meta-analysis provide evidence that the presence of the Alu insertion is not associated with an increased risk of breast cancer. Subgroup analyses by ethnicity or source of controls were used to investigate potential disequilibrium in the distribution of cases and controls. We also performed a sensitivity analysis, which reinforced the validity of the results.
Associations between PGR variation and breast cancer varied across different populations. The Alu insertion was related to breast cancer in both Indians and Indo-European mixed racial groups, while this association disappeared in Caucasians and Latinos. This difference in association is likely due to two factors. First, the various genetic backgrounds of the racial groups may contribute as the frequency of the Alu insertion varies across different populations. Certainly, genetic factors confer higher risk levels for breast cancer [41]. However, different populations also have different life-styles and are often influenced by different environmental factors [42]. In the end, the occurrence of breast cancer is determined by the interaction between genetic factors and the environment.
Although the exact biological role of the Alu insertion into the PGR gene is not yet clear, it has been reported that the insertion might cause abnormal gene transcription and weaken the binding of progesterone to the PCR, which would subsequently reduce the activity of progesterone [22]. Therefore, PGR dysfunction caused by the Alu insertion may potentially affect the occurrence of breast cancer.
Finally, there are several potential limitations of our study. First, there was measurable heterogeneity in the overall meta-analysis as well as in the sub-group analysis, which suggests that the underlying factors might partially contribute to the observed heterogeneity. Second, it is possible that the sample size was not large enough to generate a meaningful conclusion. Limited sample sizes are usually accompanied by selection bias. Thus, pooled results based on limited studies lack sufficient power to support or deny an association [43]. Third, in the ethnicity sub-group analyses, there was only one study that compared certain populations (African-American, Latino, Indian, and Indo-European) and only six studies that specified Caucasians. Thus, the discrepancy of association among different ethnic sub-groups should be interpreted cautiously. Finally, gene-gene interactions and epigenetic influences were not measured in our meta-analysis because of the limited availability of such information.
Conclusions
In conclusion, our results suggest that Alu insertion into the PGR gene is not associated with the risk of breast cancer. More studies with larger sample sizes will be needed to validate our findings and to explore potential epigenetic mechanisms and environmental influences on the risk of breast cancer.
References
Siegel R, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–41.
Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6(6):229–39.
Gomez Flores-Ramos L, et al. Association of the tumor necrosis factor-alpha -308G>a polymorphism with breast cancer in Mexican women. Genet Mol Res. 2013;12(4):5680–93.
Abdulrahman GO Jr, Rahman GA. Epidemiology of breast cancer in europe and Africa. J Cancer Epidemiol. 2012;2012:915610.
Bandera EV, et al. Racial and ethnic disparities in the impact of obesity on breast cancer risk and survival: a global perspective. Adv Nutr. 2015;6(6):803–19.
Marquina-Sanchez B et al. The interplay between intracellular progesterone receptor and PKC plays a key role in migration and invasion of human glioblastoma cells. J Steroid Biochem Mol Biol. 2017;172:198-206.
Valadez-Cosmes P, et al. Membrane progesterone receptors in reproduction and cancer. Mol Cell Endocrinol. 2016;434:166–75.
Knutson TP, et al. Phosphorylated and sumoylation-deficient progesterone receptors drive proliferative gene signatures during breast cancer progression. Breast Cancer Res. 2012;14(3):R95.
Liu D. Gene signatures of estrogen and progesterone receptor pathways predict the prognosis of colorectal cancer. FEBS J. 2016;283(16):3115–33.
Yu KD, Chen AX, Shao ZM. No association between a progesterone receptor gene promoter polymorphism (+331G>a) and breast cancer risk in Caucasian women: evidence from a literature-based meta-analysis. Breast Cancer Res Treat. 2010;122(3):853–8.
Gabriel CA, et al. Association of progesterone receptor gene (PGR) variants and breast cancer risk in African American women. Breast Cancer Res Treat. 2013;139(3):833–43.
Gaudet MM, et al. Five polymorphisms and breast cancer risk: results from the breast cancer association consortium. Cancer Epidemiol Biomark Prev. 2009;18(5):1610–6.
Haddad, S.A., et al., Hormone-related pathways and risk of breast cancer subtypes in African American women. Breast Cancer Res Treat, 2015. 154(1): p. 145-54.
Reding KW, et al. Genetic variation in the progesterone receptor and metabolism pathways and hormone therapy in relation to breast cancer risk. Am J Epidemiol. 2009;170(10):1241–9.
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703.
Zhu M, et al. AluYb8 insertion in the MUTYH gene and risk of early-onset breast and gastric cancers in the Chinese population. Asian Pac J Cancer Prev. 2011;12(6):1451–5.
Dobrovolny R, et al. Detection of large gene rearrangements in X-linked genes by dosage analysis: identification of novel alpha-galactosidase a (GLA) deletions causing Fabry disease. Hum Mutat. 2011;32(6):688–95.
Taskesen M, et al. Novel Alu retrotransposon insertion leading to Alstrom syndrome. Hum Genet. 2012;131(3):407–13.
Wada T, et al. Alu-mediated large deletion of the CDSN gene as a cause of peeling skin disease. Clin Genet. 2014;86(4):383–6.
Kim S, et al. Structural variation of Alu element and human disease. Genomics Inform. 2016;14(3):70–7.
Stenzig J, et al. Progesterone receptor variants associated with the PROGINS haplotype exhibit functional properties similar to those of wild-type progesterone receptor. Pharmacogenet Genomics. 2012;22(8):629–41.
Gallegos-Arreola MP, et al. Association of the Alu insertion polymorphism in the progesterone receptor gene with breast cancer in a Mexican population. Arch Med Sci. 2015;11(3):551–60.
Wang-Gohrke S, et al. Progesterone receptor gene polymorphism is associated with decreased risk for breast cancer by age 50. Cancer Res. 2000;60(9):2348–50.
Fabjani G, et al. Human progesterone receptor gene polymorphism PROGINS and risk for breast cancer in Austrian women. Breast Cancer Res Treat. 2002;72(2):131–7.
Lancaster JM, et al. Progesterone receptor gene polymorphism and risk for breast and ovarian cancer. Br J Cancer. 1998;78(2):277.
Donaldson CJ, et al. PROGINS Alu insertion and human genomic diversity. Mutat Res. 2002;501(1–2):137–41.
Yang B, et al. Associations of MTHFR gene polymorphisms with hypertension and hypertension in pregnancy: a meta-analysis from 114 studies with 15411 cases and 21970 controls. PLoS One. 2014;9(2):e87497.
Thakkinstian A, et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–11.
Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20(9):439–44.
Thakkinstian A, et al. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–306.
Higgins JP, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28(2):123–37.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Egger M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Yao J, et al. Association between DRD2 (rs1799732 and rs1801028) and ANKK1 (rs1800497) polymorphisms and schizophrenia: a meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(1):1–13.
Manolitsas TP, et al. No association of a 306-bp insertion polymorphism in the progesterone receptor gene with ovarian and breast cancer. Br J Cancer. 1997;75(9):1398–9.
Baracat J.L.N.S.E.N.O.F.F. Polymorphism in genes of the progesterone receptor (PROGINS) in women with breast cancer: a case-control study. Rev Bras Ginecol Obstet. 2005;27(8):473-8.
Romano A, Baars M, Martens H, Brandao R, Detisch Y, et al. Impact of Two Functional Progesterone Receptor Polymorphisms (PRP): +331G/A and PROGINS on the Cancer Risks in Familial Breast/Ovarian Cancer. Open Cancer J. 2007;1:1–8.
Surekha S, Nageswararao R. Lack of influence of progin polymorphism in breast cancer development and progression. J Cell Tissue Res. 2009;9(3):1995-8.
Arj-Ong S, et al. A systematic review and meta-analysis of tumor necrosis factor alpha-308 polymorphism and Kawasaki disease. Pediatr Int. 2010;52(4):527–32.
Moller S, et al. The heritability of breast cancer among women in the Nordic twin study of cancer. Cancer Epidemiol Biomark Prev. 2016;25(1):145–50.
Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-cancer database. Int J Cancer. 2002;99(2):260–6.
Zhong S, et al. Methionine synthase A2756G polymorphism and breast cancer risk: an up-to-date meta-analysis. Gene. 2013;527(2):510–5.
Acknowledgments
This manuscript has been edited by native English-speaking experts of BioMed Proofreading.
Funding
This study was supported by National Natural Science Foundation of China (No. 81601653).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
XLQ participated in the design of the study and performed the statistical analysis. JY carried out the studies and drafted the manuscript. YZ conceived the study and participated in its design and coordination. All authors contributed toward the drafting and revising of the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yao, J., Qi, Xl. & Zhang, Y. The Alu-insertion progesterone receptor gene polymorphism is not associated with breast cancer: a meta-analysis. BMC Med Genet 19, 16 (2018). https://doi.org/10.1186/s12881-018-0529-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-018-0529-5