Abstract
Background
Antimicrobial resistance is a global public health concern contributing to increased morbidity and mortality particularly in low-income countries. Studies on commensal bacteria are important as they reflect the state of antimicrobial susceptibility patterns in populations. However, susceptibility data on potentially pathogenic commensal bacteria from individuals in communities are still limited. The aim of this cross-sectional study was to determine the susceptibility profiles of Escherichia coli and Klebsiella species isolated from clients attending outpatient clinics in Kampala (urban district) and two rural districts of Uganda, Kayunga and Mpigi. Factors associated with such carriage are also reported.
Results
A total of 1448 participants were recruited into the study with 985 yielding organisms of interest from stool or urine samples (one per client). Most growth occurred from stool samples (636/985, 87 %), of which 620/636 (97 %) grew E. coli while 16 (3 %) were Klebsiella pneumoniae. Growth from urine was 349/985 (35 %) of which 310/349 (89 %) were E. coli while 39 (11 %) K. pneumoniae. High rates of antimicrobial resistance were detected among E. coli and Klebsiella isolates combined: sulphamethoxazole/trimethoprim 70 %, amoxicillin/clavulanate 36 %, chloramphenicol 20 %, ciprofloxacin 11 %, gentamicin 11 %, nitrofurantoin 4 %, ceftriaxone 3 %, piperacillin/tazobactam 27 %, cefoxitin 22 %, and cefepime 15 %. Multidrug resistance was noted in 33 % of the isolates. None of the isolates were resistant to imipenem. Overall, isolates from Kampala were more resistant to antimicrobials. Across the three districts combined, isolates producing beta-lactamase enzymes extended spectrum β-lactamase-(ESBL) and AmpC comprised 5.3 and 13.2 %, respectively. Further, medical procedures involving inoculation were independent risk factors [aOR 50.76 (1.80, 1432.90)] while residing in a rural district and use of sulphamethoxazole/trimethoprim 3 months prior to visiting the outpatient clinics were protective against carriage of multidrug resistant isolates. Furthermore, use of gentamicin was protective against AmpC producing isolates while clients attending HIV/AIDs clinics were less likely to carry such isolates. No factor was independently associated with carriage of ESBL-producing isolates.
Conclusion
Antimicrobial resistance is prevalent among E. coli and K. pneumoniae carried in the gut of clients attending outpatient clinics in Kampala and two rural districts in Uganda. This could complicate treatment options for community-acquired infections caused by the Enterobacteriaceae.
Similar content being viewed by others
Background
Antimicrobial resistance has become an issue of public health concern [1]; it is a major factor contributing to mortality and morbidity in settings with limited diagnostic facilities and treatment options. Fecal Escherichia coli is as an indicator of spread of acquired resistance-encoding genes in the community [2–4]. Klebsiella species on the other hand are opportunistic pathogens that play significant roles in hospital-acquired infections. Studies have shown that Klebsiella species have diverse resistance patterns to β-lactam agents. Although they are components of the normal flora, E. coli and Klebsiella species are considered potential causes of both community and hospital acquired infections [5].
Studies on commensal bacteria provide a more accurate reflection of the overall antimicrobial resistance burden in the population than data based on pathogenic isolates. In fact, studies on commensal bacteria in developing countries have shown high resistance rates to diverse antimicrobial agents [6]. In addition, drug resistance surveillance has revealed that asymptomatic carriers in the community are often colonized with resistant bacteria that subsequently lead to infection [7, 8]. Such studies are useful in the monitoring and understanding resistance mediated selection in populations [9].
In Uganda, antibiotics are readily available over the counter in community pharmacies [10], which portends significant rates of resistance among flora circulating within the community. In addition, inappropriate use of antimicrobials by pastoralists for animal diseases and the carriage of drug resistant potentially nosocomial bacteria in the livestock gut has been documented [11, 12]. Currently, most available data focuses on susceptibility of isolates from established infections without much attention on commensal isolates, particularly the Enterobacteriaceae [11, 12]. Furthermore, drug resistance is also prevalent in hospital settings in Uganda [13]. Moreover, the health care seeking behaviors in the community may vary depending on socioeconomic status of individuals involved [14]. Yet, information on resistance surveillance particularly for isolates from community settings is scarce as surveillance mainly focuses on susceptibility of isolates collected from clinical specimens in hospitals. The aim of this study was to characterize the antimicrobial susceptibility profiles of E. coli and Klebsiella species isolated as flora from faeces and urine of clients attending outpatient clinics in Uganda, and to investigate the factors associated with carriage of drug resistant isolates.
Methods
Study design
This was a cross-sectional study in which antimicrobial susceptibility testing was performed for E. coli and Klebsiella species isolated from stool or urine of asymptomatic clients attending outpatient clinics.
Study sites and setting
Uganda comprises 111 administrative districts and one city, Kampala. Districts are further subdivided into counties and sub-counties. Uganda has three types of healthcare facilities; ‘Public’, ‘Private Not for Profit’ (PNFP) and ‘Private for Profit’. The Public and PNFP facilities are the most visited by Ugandans serving about 70 % of the population [15]. The levels of healthcare together with protocols guiding treatment are described elsewhere [10].
There were two study sites categorized ‘urban’ or ‘rural’ with varied distance and district human poverty index (HPI) [16]. The study districts were purposively selected with the assumption that urban areas favor increased exposure to antimicrobials and transmission of bacteria is less in areas of low humidity compared to those of high humidity [17]. The selected urban district and one of the rural districts are of wet tropical climate [18], while the other rural district is of wet and dry tropical climate. The urban site was Kampala district, which is the capital and largest city in Uganda with a HPI of 20.5. The rural sites included two districts in central Uganda namely, Kayunga 74 km and Mpigi 29.7 km from Kampala with HPI of 30.8 and 27.5, respectively, Fig. 1. The study was conducted among individuals attending outpatient clinics at 44 Public and PNFP hospitals or health centres including 16 in Kampala, 9 in Kayunga, and 19 in Mpigi.
Study sites. The map was obtained from the URL to this figure is https://www.commons.wikimedia.org/wiki/File:Uganda_sub-counties.png, but slightly modified to emphasize the study sites
Study population and sample size estimation
The number of clients recruited from each facility was proportional to the contribution of the facility to the outpatient clinic attendance in the 3 months of April, May and June of the year 2006. The study was conducted in the second quarter of 2007 as a pilot in the urban district and the second quarter of 2008 for the two rural districts. All clients attending clinics were eligible to participate and the exclusion criteria was failure to provide either a stool or urine sample.
In Uganda no data exists on antimicrobial resistance among E. coli and/or Klebsiella pneumoniae in well-defined community infections. As such, the sample size was computed based on the prevalence of K. pneumoniae carriage in urine which has been observed to be about 19.6 % among clinical samples at clinical microbiology laboratory, College of Health Sciences, Makerere University (unpublished observations). The formula N = [(Z(α/2))2 *P *(1 − p)]/d2 = 242 where; Z = 1.96 (the standard normal deviation at 95 % CI), P = estimated prevalence of the problem under study (P = 0.196), d = margin of error (precision) of prevalence estimate at 5 % (0.05), α = level of significance at 5 % and n = required sample size was used. Primary sampling clusters were used to recruit individuals and thus, using a design effect of 2.0, the sample size under cluster sampling was 484 per district and 1452 for the three districts combined.
Selection of health care facilities and participants
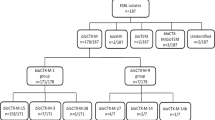
In each of the three districts multistage sampling was done based on the average clinic attendance for each district to ensure a sample size of 484, given an observed frequency of Klebsiella species in 19.6 % of urine specimens and a design effect of 2, Fig. 2. Thirty clusters of 16–20 participants were selected from each district using the probability proportion to size sampling, which ensured all individuals in the target population had an equal chance of being selected. Two busy days of a week were purposively selected to visit each health care facility. When the number exceeded 20, systematic sampling was carried out.
Data collection
A standardized interviewer administered questionnaire (Additional file 1: Figure S1) was used to collect data on socio-demographics (age, sex, and residence), heath care unit, and reason for visit and history of use of antimicrobial agents, history of admission and history of medical procedures in the previous 3 months.
Sample collection
Participants were instructed to provide a stool or a urine sample if they were unable to provide stool. The specimens were collected in sterile containers with screw caps. Samples from rural districts were stored at 4 °C for one to 3 days prior to transportation by road in cool boxes while those from Kampala were transported to the laboratory on the day of collection.
Cultures
Isolation of E. coli and Klebsiella species and the antimicrobial susceptibility testing were performed at the Clinical Microbiology Laboratory, College of Health Sciences, Makerere University. The laboratory participates in the College of American Pathologist bacteriology external quality assurance scheme (CAP no. 7225593).
Samples were streaked onto MacConkey agar and incubated for 18–24 h at 37 °C in ambient air. In case of stool, specimens were first emulsified in sterile normal saline before inoculation of the MacConkey plates. Lactose fermenting isolates, from the fourth or third streak area with colonial morphologies suggestive of E. coli and Klebsiella species were subjected to an oxidase test, and when negative, they were cultured for 18–24 h on the following media: triple sugar and iron (TSI) agar, Simmon’s citrate agar, and sulphide-indole-motility (SIM) media for identification. Following identification, E. coli and Klebsiella isolates with inconclusive identification were confirmed with the API 20E system (BioMerieux Marcy 1’Etoile, France).
Antimicrobial susceptibility testing
Drug susceptibility testing (DST) was performed with the disc diffusion method on Mueller Hinton Agar (MHA) (Biolab, Hungary) plates as recommended by the clinical laboratory standards institute (CSLI). Three colonies were emulsified into sterile saline and the turbidity of the suspension adjusted to the 0.5 McFarland standard. The antimicrobial disks (Biolab, Hungary) used included ampicillin (10 µg), amoxicillin/clavulanate (20/10 µg), cefuroxime (30 µg), ceftriaxone (30 µg), cefotaxime (30 µg), ceftazidime (30 µg), meropenem (30 µg), trimethoprim/sulfamethoxazole (1.25/23.75 µg), chloramphenicol (30 µg), gentamicin (10 µg), ciprofloxacin (5 µg), nitrofurantoin (300 µg), cefepime (30 µg), piperacillin/tazobactam (100/10 µg) and cefoxitin (30 µg).
Escherichia coli ATCC 25992/ATCC 35218, Staphylococcus aureus ATCC 25923, and Pseudomonas aeruginosa ATCC 27853 were used to quality control the susceptibility testing procedures in line with CLSI guidelines.
For isolates resistant to second or third generation cephalosporins, minimum inhibitory concentrations (MICs) were performed by E-test (AB BIODISK Solna, Sweden) according to the manufacturer’s instructions and these were confirmed using the Vitek 2 identification and susceptibility system (Biometric, Marcy I’Etoile, France). The results were interpreted according to CLSI guidelines [19].
Detection of extended spectrum β-lactamase production
Detection of extended spectrum β-lactamases (ESBLs) was performed using the double disc synergy test (DDST) using amoxicillin/clavulanate, ceftazidime, ceftriaxone, aztreonam, and cefotaxime as previously described [20, 21]. E-test strips (containing cefotaxime/ceftotaxime-clavulanate, ceftazidime/ceftazidime-clavulanate and cefepime/cefepime-clavulanate) (AB BIODISC, Solna, Sweden) were used. K. pneumoniae 70,063 was used to control ESBL detection. The E test ESBL strip (AB Biodisk, Solna, Sweden) carries two gradients: on one end, ceftazidime or cefotaxime; and on the opposite end, ceftazidime or cefotaxime plus clavulanic acid. MIC were interpreted as the point of intersection of the inhibition ellipse with the E test strip edge. A ratio of ceftazidime or cefotaxime MIC to ceftazidime or cefotaxime-clavulanic acid MIC equal to or greater than eight indicated the presence of ESBL (manufacturer’s instructions) [22].
Phenotypic detection of AmpC enzymes
Phenotypic detection of AmpC β-lactamase production was performed on isolates with reduced cefoxitin diameters (≤17 mm). These were sub-cultured on MHA containing 8 μg/ml of cefoxitin agar (USP reference standard, Rockville) and screened with E-test strips (cefotetan/cefotetan-cloxacillin, CN/CNI AB BIODISC, Solna, Sweden). A ratio of ≥8, deformation of the ellipse, or the presence of a phantom zone were interpreted as positive for AmpC production [23].
Data analysis
Questionnaire responses and DST data were checked for completeness. Data was double entered for validation using EPIDATA software, cleaned and exported to STATA (v10) for analysis. Data were analyzed using descriptive statistics, frequencies and bivariate analyses (cross-tabulations). Associations were tested using Pearson’s Chi square. A significant level was set at p = 0.05. Odds ratios (OR) were determined between socio-demographic variables, health facility level, district, reason for visit, prior use of antimicrobial, history of hospital admission and medical procedures 3 months prior to visits, with the following outcomes; (1) resistance to one antibiotic, (2) resistance to three different classes of antimicrobial agents -multi-drug resistance (MDR), (3) ESBL phenotype, (4) AmpC phenotype.
Variables with p < 0.15 were entered into multivariate logistic regression models with backward elimination. To control for the effect of clustering, regression with robust standard errors was used. Independent variables used were; sex (male versus female), health center level, health sub-district and district, history of admission, procedures and antibiotic use recalled by client and from health record, in the previous 3 months.
Differences between proportions of resistant isolates were determined to compare variation within districts using MDR as surrogate marker. MDR was determined based on categories and drugs used to determine susceptibility among the Enterobacteriaceae [24]. The categories included: aminoglycosides (gentamicin); anti-pseudomonal penicillin/β-lactamase inhibitor (piperacillin/tazobactam); carbapenem (meropenem); non-extended spectrum cephalosporins (cefuroxime); extended spectrum cephalosporin (ceftriaxone, ceftazidime, cefotaxime, cefepime); cephamycins (cefoxitin); fluoroquinolone (ciprofloxacin); folate pathway inhibitors (sulphamethoxazole/trimethoprim); penicillins (ampicillin); and phenicols (chloramphenicol). These 11 antimicrobial agents represented 11 of the 16 recommended categories however, second and third generation cephalosporins were considered as oxyminocephalosporins.
Results
A total of 1448 participants were recruited; 474 (33 %) Kampala, 508 (35 %) Kayunga, and 466 (32 %) Mpigi. Females were 913 (63.3 %) while men were 529 (36.7 %) and the difference was statistically significant (p = 0.005). The mean age of the participants was 24.8 years (SD 17.5) (Table 1); 802 (55.7 %) of them belonged to the 15–44 year age group. Participants provided 730 and 718 stool and urine samples, respectively, which were cultured for identification of E. coli and Klebsiella isolates.
Socio-demographic factors of participants
Five hundred and thirty-five participants visited the clinics for general conditions and these comprised headache and fever (422, 28.49 %); follow-up review (85, 5.74 %); antenatal care 20 (1.35 %); and cardiovascular diseases (15, 1.01 %). Other reasons for visits included infections (731, 52.7 %), such as respiratory tract infections (312, 21.07 %); gastro-intestinal (309, 20.86 %); genital urinary infections (125, 8.44 %); skin infections (64, 4.32 %); and attending HIV/AIDS clinics (122, 8.71 %). Use of antimicrobials 3 months prior to the study was high (1093, 75.5 %); septrin (774, 70.6 %); penicillins (369, 33.9 %), ciprofloxacin (130, 11.9 %); and gentamicin (45, 4.1 %), Table 1. One hundred and twenty-five (125, 8.7 %) participants had been admitted to hospital 3 months prior to the clinic visit while 130 (8.9 %) underwent medical procedures, of which 17 (13.1 %) involved only contact (such as radiological investigations) while 88 (67.7 %) had investigations like taking blood for malaria, injections and infusion (inoculation), and 25 (19.2 %) had minor or major surgery. Overall, there was significant variation among the parameters related with resistance across the three districts, Table 1.
Prevalence of E. coli and Klebsiella species
Of the 1448 participants, growth of E. coli or K. pneumoniae was obtained from 985 samples and significant variation was observed across the three districts; 274 Kampala, 406 Kayunga, and 305 Mpigi (p < 0.001). Growth from stool occurred in 636 (87 %) samples of which 620 (97 %) were E. coli and 16 (3 %) were K. pneumoniae. Growth from urine occurred in 349 (49 %) samples of which 310 (89 %) were E. coli and 39 (11 %) K. pneumoniae; none of the urine samples had growth of ≥104 colony forming units (CFU) implying there was no infection-related growth. Forty-one samples grew both E. coli and K. pneumoniae; where both grew, E. coli was considered as it is the commonest cause of community infections. Overall 930 (94.4 %) E. coli and 55 (5.6 %) K. pneumoniae isolates were isolated and studied.
Susceptibility profiles
Overall, two hundred and seven (207, 21 %) isolates were susceptible to all antimicrobial agents tested;
-
1.
β-lactam agents; 617 (67 %) E. coli isolates were resistant to ampicillin (Klebsiella were omitted as they are inherently resistant to ampicillin). Resistance to other drugs was (for both Klebsiella and E. coli); 145 (14 %) cefuroxime, 29 (3 %) ceftriaxone, and 359 (36 %) amoxicillin/clavulanate. Further analysis revealed resistance in 263 (27 %) isolates for piperacillin/tazobactam, 213 (22 %) cefoxitin and 152 (15 %) cefepime, Table 2. Although none of the isolates was resistant to meropenem, zone diameters of ≤ 25 mm were noted in 167 (17 %) of the isolates a finding that necessitates screening for carbapenamase activity.
Table 2 Antimicrobial resistance profiles of E. coli and Klebsiella pneumoniae in the three districts -
2.
Other antimicrobial agents. High resistance against sulphamethoxazole/trimethoprim (698, 69 %), chloramphenicol (19 %, 195), ciprofloxacin (107, 11 %), gentamicin (106, 11 %), and nitrofurantoin (40, 4 %) was detected. Overall, resistance did not vary by gender except that isolates from females 123/167 (74 %) were more likely to have reduced susceptibility (inhibition zone diameter ≤25) to carbapenems (p < 0.023).
-
3.
MDR was noted in 356 (36.1 %) isolates and there was significant variation across districts (p < 0.001). Three hundred and twenty-three E. coli isolates (323/930, 34.7 %) and 33 (33/55, 60 %) Klebsiella isolates were MDR and the finding was statistically significant (p < 0.001). The highest MDR prevalence was observed in Kampala, an urban district. MDR phenotype also varied by health sub-districts (HSD) ranging from 9/46 (19.6 %) in rural to 26/33 (79 %) in urban. Further, the MDR phenotype was detected in all health care facilities in Kayunga (9) and Kampala (16), and was not detected in 4 of the 19 health care facilities in Mpigi. Among the Ampicillin resistant isolates, 32 (3 %) were resistant to nitrofurantoin, 94 (9 %) gentamicin, 100 (10 %) ciprofloxacin, 192 (19 %) chloramphenicol and 592 (60 %) septrin. In addition, there was high resistance among the isolates to multiple drug combinations, Table 3.
Table 3 Frequency and patterns of resistance phenotypes among isolates from the study sites
Prevalence of ESBL-producing isolates
Overall, 209 bacterial isolates from 40 health care centers qualified for AmpC screening (based on cefoxitin resistance) while 63 isolates from 23 health centers qualified for ESBL-screening (based on CLSI guidelines). AmpC and ESBL-producing isolates were higher in Kampala [ESBL; 35/274 (12.8 %), AmpC; 65/274 (23.7 %)] compared to Kayunga [ESBL 7/406 (1.7 %), AmpC; 27/406 (6.7 %)] and Mpigi [10/305 (3.3 %), AmpC; 37/305 (12.1 %)]. However, among isolates that qualified for screening for ESBLs, there was no significant difference in the distribution of the enzymes across the three districts i.e. for ESBLs in 35 (83.3 %), 7 (77.8 %) and 10 (83.3 %) isolates (p = 0.886) and AmpC in 65 (53.3 %), 27 (46.6 %) and 37 (55.2 %) isolates (p = 0.505); for both enzymes in Kampala, Kayunga and Mpigi, respectively.
AmpC enzymes were detected in 61 % (116/191) and 72 % (13/18) E. coli and Klebsiella, respectively (χ2 (1) 4.9527, p = 0.026). ESBLs were detected in 81 % (42/52) E. coli and 100 % (10/10) of the screened Klebsiella isolates. The coexistence of ESBLs and AmpC enzymes was detected in 28.8 % (15/52) of the isolates. The cefoxitin inhibition zone diameters of ≤13 mm were significantly associated with presence of AmpC enzymes in both E. coli and Klebsiella isolates (p < 0.038) but it was not the case for ESBLs when reduced zone diameters recommended for ESBL screen (CLSI) were used (p = 1.00).
The overall prevalence of AmpC phenotype was 13 % (129/985) for the three districts. Whereas 97 of the 182 MDR screened had AmpC enzyme, this was not statistically significant [χ2 (1) = 0.0253, p = 0.874]. Isolates bearing AmpC β-lactamase were sensitive to gentamicin 96/129 (74 %) [χ2 (1) 4.766, p < 0.029] while 74 % (90/122) of them were resistant to piperacillin/tazobactam much as this association was not significant [χ2 (1) 0.766, p = 0.381].
The prevalence of the ESBL phenotype was 5.3 % (52/985). Among isolates bearing ESBLs β-lactamase and were resistant to amoxicillin/clavulanate, 86 % (32/37) were also resistant to piperacillin/tazobactam [χ2 (1) 0.087, p = 0.767].
Univariate analysis and logistic regression model
On univariate analysis, significant risk factors for carriage of ESBLs among outpatient clinic attendees were Health center level (p = 0.037) and use of antibacterial agent in the last 3 months prior to clinic visit (p = 0.023), Additional file 2: Table S1. In addition to these the health sub-districts with less than 100 participants with growth (p = 0.079) and use of septrin (p = 0.088) were the variables included in the logistic regression model. Using backward stepwise procedure, no independent risk factor for carriage of ESBL enzymes was identified. However, without controlling (for other factors), use of antimicrobials 3 months prior to clinic visit was found to be associated with carriage of ESBL cOR 4.47 (95 % CI 1.15, 17.44), Table 4.
Significant risk factors at univariate analysis for carriage of AmpC enzymes were reason for coming to clinic (p = 0.003), history of admission (p = 0.023) and history of medical procedures (p = 0.044), Additional file 2: Table S1. These variables, together with sex (p = 0.132) and use of gentamicin (p = 0.100) were included in the model. Following backward stepwise procedure, visiting the outpatient clinic for reasons other than infection or HIV/AIDs care [aOR 3.56 (95 % CI 1.08, 11.77)] was an independent risk factor for carriage of AmpC producing E. coli and K. pneumoniae while use of gentamicin was independent of such carriage [aOR 0.17 (95 % CI 0.03–0.95]. However, without controlling for other factors, history of admission was found to be associated with carriage of AmpC [cOR 2.76 (95 % CI 1.12, 6.84)], Table 4.
MDR isolates were prevalent across the three districts and in the univariate analysis the significant factors for carriage of MDR isolates were age group (p = 0.004), health center level (p < 0.001), health sub-district (p < 0.001), district of residence (p < 0.001), health sub-district (group of <100 participants with growth (p < 0.001), Additional file 2: Table S1, and use of septrin (p < 0.001), Additional file 2: Table S1 (Additional file 3: Table S2). In multivariate analysis four variables; age group, health center level, district of residence, and use of septrin together with history of medical procedures (p = 0.075), use of antimicrobial agents (p = 0.122); including use of ciprofloxacin (p = 0.071) and were included in the model. Using backward stepwise procedure, having had medical procedures involving inoculation in the last 3 months prior to visiting health facility was independent risk factor for carriage of MDR isolates [aOR 50.76 (95 % CI 1.80–1432.90)]. However, without controlling for other factors when compared to residence in Kampala, residing in Kayunga and Mpigi was protective by 83 % (95 % CI 0.11–0.26) and 80 % (95 % CI 0.13, 0.32), respectively and septrin use was protective by 47 % (95 % CI 0.40, 0.71) of carriage of MDR E. coli or K. pneumoniae. Furthermore, the age group 15–44 was 1.65 times more likely to have MDR isolates (95 % CI 1.22, 2.23) compared to participants less than 15 years and HSD with less than 100 cases were 1.75 times more likely to have such isolates (0.5 % CI 1.33–2.29). Also, attending lower hospitals was protective from carriage of MDR isolates when compared to National referral hospitals [cOR 0.32 (95 % CI 0.20, 0.52)] for general hospitals, HCIV [cOR 0.27 (95 % CI 0.16, 0.46)] and HCIII [cOR 0.21 (95 % CI 0.13, 0.33)], Table 4.
Discussion
We have reported the prevalence and susceptibility patterns of E. coli and K. pneumoniae isolated from the intestinal and urinary tracts of individuals attending urban and rural outpatient clinics in Uganda. In order to study susceptibility of flora among outpatient department (OPD) clients for the first time, the study was conducted in two phases a pilot study that took place between May and June of 2007, and involved generating sampling frame, seeking permission and recruitment took place in Kampala district. The aim was to test the recruitment strategy and sample collection, transportation and analysis. The experience from the pilot study was then used for study activities in the two rural districts.
The findings in this study reveal high rates of resistance to commonly used antibiotics such as ampicillin and septrin and relatively lower resistance rates to amoxicillin/clavulanate, chloramphenicol, ciprofloxacin, gentamicin, nitrofurantoin and ceftriaxone. These findings are similar to those from neighboring Kenya [25]. The high levels of resistance to piperacillin/tazobactam and cefoxitin among the amoxicillin/clavulanate resistant isolates found were associated with AmpC enzymes. Despite the low resistance to third generation cephalosporins, ESBLs were detected in 5.3 % isolates most of which were MDR with co-resistance to ciprofloxacin, septrin and gentamicin. However, the MDR rates in our study was lower than that of fecal carriage among outpatient department volunteers in Cameroon [26]. This study also found that E. coli is the species most commonly isolated among ESBL producers [26]. The prevalence of ESBLs in our study was similar to findings among French Guiana Amerindians living in a very remote village [27]. There was significant variability among the three districts, one possible reason could be the nature of samples provided, when individuals visit outpatient clinics, the convenient specimen depends on the respective toilet habits of participants and possibly diet, however the nature of bathrooms at the clinic may not be conducive for a participant to take much time there in. In Uganda communities with the wet and dry climate tend to have diets different from those of wet climate. The variation in sex distribution cannot be explained, neither is the presence of more elderly people in the rural district of Mpigi, this same district prescribed more ciprofloxacin, and gentamicin, had higher admissions and surgeries too.
Susceptibility patterns
The resistance to antibacterial agents tested significantly varied across the three districts and was highest in the urban district of Kampala, save for cefoxitin where it was higher in the western rural district of Mpigi. Resistance to piperacillin/tazobactam did not significantly vary across the districts but was highest in the eastern rural district of Kayunga. Our findings are consistent with previous studies that reported higher resistance in urban compared to rural areas [28, 29]. Resistance also varied within districts possibly reflecting differences in services offered by health care givers. It is also possible that segregation in community activities where some are entirely agricultural based with sparse population lessens access to antibiotics. Populations having commercial activity and higher population density create opportunity for increased environmental density of antibiotic use and antibiotic resistant bacteria as a result of access to drug outlets and health facilities [28].
In this study, over 76 % of participants reported prior use of antimicrobial agents particularly septrin and penicillins. The prevalence of HIV in Uganda necessitates use of antimicrobials for treatment of opportunistic infections, and the routine use of septrin prophylaxis against Pneumocystis jirovecii pneumonia. This could contributes to bacterial resistance [30, 31]. However in this study the use of septrin was protective against carriage MDR E. coli and K. pneumoniae. This finding requires focused studies to determine factors underlying this protection. Furthermore, individuals with HIV/AIDs were significantly less likely to have E. coli and K. pneumoniae producing AmpC enzymes; this may be attributed to the decrease in infections attributed to use of basic care package (BCP), a patient managed, home based care system introduced since 2005 and is widely used [32]. It empowers HIV-positive people to avert opportunistic infections and exposure to malaria transmitting mosquito among many other benefits; thus living a healthier life and preventing occurrence of diseases [33, 34]. This possibly keeps these individuals away from frequent exposure to health facilities, a factor likely to be contributing to carriage of AmpC in this study, and visiting drug outlets thus indirectly protecting them from exposure to AmpC-bearing Enterobacteriaceae. There was little use of third generation cephalosporins, β-lactamase inhibitors and cephamycins and yet resistance to the respective classes was detected among study participants. There is a possibility that use of antimicrobial agents to which bacteria are already resistant selects for further resistance and also co-selects resistant determinants that may be co linked to resistance of the respective drug in use.
The resistance to cefepime was noted to be higher than that of ceftriaxone, such resistance may be due to presence of oxacillinase enzymes like Oxa-30 and Oxa-31, one of the derivatives of Oxa-1, that confers a spectrum sparing ceftazidime and cefotaxime and hydrolyses cefepime, but it has also been reported to be associated with alteration or loss of outer membrane porins [35, 36]; these possibilities should be explored in subsequent studies.
The commonest MDR combination when associated with beta-lactamase enzymes determinants are likely to be selected for by use of respective antimicrobials, thus increasing the magnitude of beta-lactamases as postulated elsewhere [37]. The health centers studied were assumed to serve over 70 % of the population. However there is inadequate laboratory diagnosis in most of the rural district areas often leaving resistance to go undetected. Under these circumstances, empiric as opposed to targeted therapy prevails [38]. In the period of the study, visits to health facilities depended on availability of medicines and stock outs were frequent consistent with findings from previous studies [39]. When the antimicrobials were not present, clients may have resorted to drug outlets [40] where doses offered are suboptimal [10] and such exposure is an established driver of resistance. Frequent use of antimicrobial creates a pool of resistant commensal bacteria that contribute to general increase and dissemination of bacterial resistance worldwide [41]. In this setting, this is compounded by the liberal use of antimicrobials by farmers [12]. Evidence from settings where antibacterial agents are accessed by prescription only, have shown that individuals prescribed an antibiotic in primary care develop bacterial resistance to the respective antibiotic, with effect being greatest in the month immediately after treatment but persistence may go on for over 12 months [42]. The high presence of septrin resistance in a setting where it is continuously used, may be explained by several of the above factors.
Factors associated with drug resistance
Multidrug resistance was common and factors such as age, level of health facility, location of health sub-district, district of residence, undergoing medical procedures and use of septrin were associated with multidrug resistance. However, only undergoing medical procedures associated with inoculation was an identified risk factor for carriage. Other factors yet to be identified contribute to the use of septrin within 3 months prior to visiting the clinics [cOR 0.53 (95 % CI 0.40, 0.71)] and district of residence whereby those from rural districts of Kayunga [cOR 0.17 (95 % CI 0.11, 0.26)] and Mpigi [cOR 0.20 (95 % CI 0.13, 0.32)] were protected from carriage of MDR flora compared to those who reside in Kampala. Furthermore visiting lower health centers like general hospital [cOR 0.32 (95 % CI 0.20, 0.52)], HCIV [cOR 0.27 (95 % CI 0.16, 0.46] and HCIII [cOR 0.21 (95 % CI 0.13, 0.33)] compared to national referral hospital was protective of MDR carriage, the age group 15–44 was a risk factor for carriage of MDRisolates compared to participants ≤15 years [cOR 1.65 (95 % CI 1.22, 2.23)] and so were HSD of less than 100 cases cOR 1.75 (95 % CI 1.33, 2.29).
Multidrug resistance was noted for over 60 % of ESBLs and 70 % AmpC bearing isolates; whereas no factor was independently associated with carriage of isolates producing ESBLs, having used antibacterial agents during 3 months prior to OPD visit may have combined with yet unknown factors to contribute to carriage of flora with the enzyme [cOR 4.47 (95 % CI 1.15, 17.44)]. Previously studies have reported that antibiotic exposure affects ESBL faecal carriage in the community [27].
The isolates with AmpC enzymes were independently associated with clinic visits for reasons other than infection or HIV/AIDs care [aOR 4.38 (95 % CI 1.14, 16.84)] while use of gentamicin was protective of such carriage [aOR 0.17 (95 % CI 0.03, 0.95)]. Participants with history of admission 3 months prior to admission were also likely to have carriage of isolates with AmpC enzymes [cOR = 2.76 (95 % CI 1.12, 6.84)]. Such findings highlight the role played by direct interaction of clients with facility environment including health care givers. The association of hospital admission and care with AmpC and MDR isolates can be explained by the levels of overcrowding in health facilities compounded by substandard infection control practice in the study setting where hand hygiene is seen more as self-protection by health care givers as opposed to breaking the chain of transmission in the facility [43]. Such practice facilitate transmission of bacteria between clients and care givers [44]. This argument is strengthened by finding procedures associated with inoculation, where direct contact with health care providers is inevitable, as an independent risk factor.
Previous studies have pointed out the acquisition of AmpC enzyme bearing bacteria from health facilities. However the presence of these enzymes in the community following the introduction by those who visited health facilities is worrying [45] as they drive the use of Carbapenems for community-acquired infections [46] and are associated with poor outcome [46].The presence of MDR isolates in varied combination some covering over five different class drugs may be an indicator of evolving gene pools in flora from the study areas [47]. In this study age and sex were not independently associated with resistance.
Population exposure to antibiotics is related to ESBL community carriage rates [27]. However several studies in the past [28] and a recent one from Australia demonstrated presence of resistance among individuals who have not taken antibiotics [48]. The presence of ESBL enzymes and associated MDR among individuals reporting to health centers without associated factors has been highlighted in other studies [49], and several sources for ESBL transmission in the community have been identified. For example ESBLs was detected from food of animal origin such as raw meat similar to faecal samples from hospitals in The Netherlands [50]. Extended-spectrum β-lactamase genes of E. coli was detected in chicken meat and humans, in the Netherlands [50]. Similarly in Uganda the resistance of E. coli isolates from faecal samples of chicken and pigs support the possibility of having resistance arising from the food chain [11]. Transmission of bacteria between human and animal sources has been reported from communities associating with wild life in Uganda [51] and in Portugal Enterobacteriaceae from health humans had resistance determinants similar to those from food producing animals [52]. Elsewhere in Kenya companion animals of Pastoralists were found to carry ESBls [53] and there is a possibility that similar scenarios of transmission of resistance take place in this setting.
The environment has been suggested to serve as a reservoir for ESBL genes since isolates from varied sources are noted to be similar to those of humans [54]. A variety of factors such as community hygiene levels, people’s behavior, and antimicrobial resistance rates at community level influence on transmission of resistant microorganisms. The transmission of commensal bacteria between individuals and antibiotic use occur all the time, this ensures selection of resistant bacteria in the community [55]. Uganda being a low resource country, several factors are likely to be playing a role in the transmission of these resistance traits. The findings from rural central Uganda show that families with poor hygienic practices and environmental hygiene were significantly associated with having helminthes transmitted by oral faecal spread, which would equally have transmission of resistant bacteria [56]. The sanitation standards in the urban areas are worsened by low levels of funding and supervision of sewage systems with frequent over flow of sewerage onto the surface [57]. On the other hand 70 % of the urban poor use shared latrines that quite often fail to serve the desired need of improved sanitation [58]. Furthermore there are unhygienic practice by food vendors in the region covering the study area [59]. In a low resource setting purchase transaction involves use of bank notes and yet in the recent past contaminated bank notes have been identified as contributing to the transmission of ESBLs [60]. It is possible this occurs among individuals who may have no history of using antibacterial agents in this setting too.
The level of resistance noted in this study has several implications. With minimal laboratory support for culture and sensitivity the chances of ineffective chemotherapy, treatment failure and increased duration of hospital stay and death are likely to be rampant [61]. With prolonged illness, there is increased risk of spread of infection to other people with ultimate increase in costs of health care [62]. Uganda being a low income country, treatment of multidrug resistant bacteria necessitating the purchase of expensive last line antimicrobials is not easy to achieve in the public facilities [63, 64]. Thus these findings call for initiation of efforts to continuously collect susceptibility data from both rural and urban areas to inform the planning of antibiotic policies and introducing antibiotic stewardship programs at all levels of health care delivery.
One limitation in this study was that we considered three colonies from all specimen with successful growth, instead of using antibiotic containing media to screen for antimicrobial resistance phenotypes. This could have led to missing some ESBLs. However, the study gives insight into the problem of MDR among ESBL negative bacteria. Use of client prescription notebooks and recall to give information about antimicrobials previously taken, may have left some antimicrobial use undetected. The use of specimen as opposed to rectal swabs affected the yield of flora. The findings from this study are based on clients attending outpatient clinics in the selected districts of central Uganda and may not be generalized to clients attending outpatient clinics throughout the country.
Conclusions
Antimicrobial resistance is wide-spread among E. coli and K. pneumoniae in clients attending outpatient clinics in Kampala and rural districts of Uganda. This could complicate treatment options for community acquired infections caused by the enterobacteriaceae.
Abbreviations
- AmpC:
-
class C beta-lactamase enzyme (cephalosporinase)
- ESBL:
-
extended spectrum beta-lacatamase enzyme
- MDR:
-
multidrug resistant
- HIV/AIDS:
-
human immunodeficiency virus/aquired immunodeficiency syndorme
- HC:
-
health center
- HSD:
-
health sub-district
- PNFP:
-
private not for profit
- HPI:
-
human poverty index
- CI:
-
confidence interval
- ISS:
-
immunosuppresive syndrome
- CAP:
-
College of American pathologists
- CLSI:
-
clinical laboratory standards institute
- DST:
-
drug susceptibility testing
- MHA:
-
Mueller–hinton agar
- SIN:
-
sulphide indo-motility agar
- TSI:
-
tripple sugra and iron
- ATCC:
-
American type culture collection
- MIC:
-
minimum inhibitory concentraion
- DDST:
-
double disk synergy test
- SD:
-
standard deviation
- IRB:
-
institutional review board
- cOR:
-
crude odds ratio
- aOR:
-
adjusted odds ratio
- BCP:
-
basic care package
- SIDA:
-
Swedish International Development Cooperation Agency
References
Gootz TD. The global problem of antibiotic resistance. Crit Rev Immunol. 2010;30(1):79–93.
Lester SC, del Pilar Pl M, Wang F, Perez Schael I, Jiang H, O’Brien TF. The carriage of Escherichia coli resistant to antimicrobial agents by healthy children in Boston, in Caracas, Venezuela, and in Qin Pu, China. N Engl J Med. 1990;323(5):285–9.
Aarestrup FM, Oliver Duran C, Burch DG. Antimicrobial resistance in swine production. Anim Health Res Rev. 2008;9(2):135–48.
Nys S, Okeke IN, Kariuki S, Dinant GJ, Driessen C, Stobberingh EE. Antibiotic resistance of faecal Escherichia coli from healthy volunteers from eight developing countries. J Antimicrob Chemother. 2004;54(5):952–5.
Hanson ND, Moland ES, Hong SG, Propst K, Novak DJ, Cavalieri SJ. Surveillance of Community-Based Reservoirs Reveals the Presence of CTX-M, Imported AmpC, and OXA-30 β-Lactamases in Urine Isolates of Klebsiella pneumoniae and Escherichia coli in a U.S. Community. Antimicrob Agents Chemother. 2008;52(10):3814–6.
Bartoloni A, Pallecchi L, Benedetti M, Fernandez C, Vallejos Y, Guzman E, Villagran AL, Mantella A, Lucchetti C, Bartalesi F. Multidrug-resistant commensal Escherichia coli in children, Peru and Bolivia. Emerg Infect Dis. 2006;12(6):907–13.
Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis. 2006;43(Supplement 2):S62–9.
Reddy P, Malczynski M, Obias A, Reiner S, Jin N, Huang J, Noskin GA, Zembower T. Screening for extended-spectrum beta-lactamase-producing Enterobacteriaceae among high-risk patients and rates of subsequent bacteremia. Clin Infect Dis. 2007;45(7):846–52.
Caprioli A, Busani L, Martel JL, Helmuth R. Monitoring of antibiotic resistance in bacteria of animal origin: epidemiological and microbiological methodologies. Int J Antimicrob Agents. 2000;14(4):295–301.
Mukonzo JK, Namuwenge PM, Okure G, Mwesige B, Namusisi OK, Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. J Multidiscip Healthc. 2013;6:303–10.
Byarugaba DK, Kisame R, Olet S. Multi-drug resistance in commensal bacteria of food of animal origin in Uganda. Afr J Microbiol Res. 2011;5(12):1539–48.
Byarugaba DK. A view on antimicrobial resistance in developing countries and responsible risk factors. Int J Antimicrob Agents. 2004;24(2):105–10.
Seni J, Najjuka CF, Kateete DP, Makobore P, Joloba ML, Kajumbula H, Kapesa A, Bwanga F. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Uganda. BMC Res Notes. 2013;6:298.
Schellenberg JA, Victora CG, Mushi A, de Savigny D, Schellenberg D, Mshinda H, Bryce J. Inequities among the very poor: health care for children in rural southern Tanzania. Lancet. 2003;361(9357):561–6.
Uganda Go: National house hold survey. Entebbe: Statistics UBo; 2000.
UNDP. Uganda human development report. Kampala: United Nations Development Programme; 2005.
Scott E, Bloomfield SF. The survival and transfer of microbial contamination via cloths, hands and utensils. J Appl Bacteriol. 1990;68(3):271–8.
Pidwirny M. Fundamentals of physical geography; 2006. In: On the web at.
CLSI. Performance standards. In: Institute CaLS, editior. M100-S17. vol. ISBN 1-56238-625-5, 1 edn. 940 West Valley Road, Suite 1400, Wayne 19087-1898 USA; 2007.
Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14(Suppl 1):90–103.
Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867–78.
Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14:90–103.
Polsfuss S, Bloemberg GV, Giger J, Meyer V, Böttger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2011;49(8):2798–803.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Kiiru J, Kariuki S, Goddeeris BM, Butaye P. Analysis of beta-lactamase phenotypes and carriage of selected beta-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol. 2012;12:155.
Lonchel CM, Meex C, Gangoue-Pieboji J, Boreux R, Assoumou MC, Melin P, De Mol P. Proportion of extended-spectrum ss-lactamase-producing Enterobacteriaceae in community setting in Ngaoundere, Cameroon. BMC Infect Dis. 2012;12:53.
Woerther PL, Angebault C, Jacquier H, Clermont O, El Mniai A, Moreau B, Djossou F, Peroz G, Catzeflis F, Denamur E, et al. Characterization of fecal extended-spectrum-beta-lactamase-producing Escherichia coli in a remote community during a long time period. Antimicrob Agents Chemother. 2013;57(10):5060–6.
Walson JL, Marshall B, Pokhrel BM, Kafle KK, Levy SB. Carriage of antibiotic-resistant fecal bacteria in Nepal reflects proximity to Kathmandu. J Infect Dis. 2001;184(9):1163–9.
Blomberg B, Olsen BE, Hinderaker SG, Langeland N, Gasheka P, Jureen R, Kvåle G, Midtvedt T. Antimicrobial resistance in urinary bacterial isolates from pregnant women in rural Tanzania: implications for public health. Scand J Infect Dis. 2005;37(4):262–8.
Wininger David A, Fass RJ. Impact of trimethoprim-sulphamethoxazole propylaxis on etiology and susceptibilities of pathogens causing human immunodeficiency virus-associated bacteremia. Antimicrob Agents Chemother. 2002;48(2):594–7.
Madhi SA, Petersen K, Madhi A, Khoosal M, Klugman KP. Increased disease burden and antibiotic resistance of bacteria causing severe community-acquired lower respiratory tract infections in human immunodeficiency virus type 1-infected children. Clin Infect Dis. 2000;31(1):170–6.
Colindres P, Mermin J, Ezati E, Kambabazi S, Buyungo P, Sekabembe L, Baryarama F, Kitabire F, Mukasa S, Kizito F, et al. Utilization of a basic care and prevention package by HIV-infected persons in Uganda. AIDS Care. 2008;20(2):139–45.
Penfold S, Simms V, Downing J, Powell RA, Mwangi-Powell F, Namisango E, Moreland S, Atieno M, Gikaara N, Kataike J. The HIV basic care package: where is it available and who receives it? Findings from a mixed methods evaluation in Kenya and Uganda. AIDS Care. 2014(ahead-of-print):1–9.
Peletz R, Simunyama M, Sarenje K, Baisley K, Filteau S, Kelly P, Clasen T. Assessing water filtration and safe storage in households with young children of HIV-positive mothers: a randomized, controlled trial in Zambia. PLoS ONE. 2012;7(10):e46548.
Dubois V, Arpin C, Quentin C, Texier-Maugein J, Poirel L, Nordmann P. Decreased susceptibility to cefepime in a clinical strain of Escherichia coli related to plasmid- and integron-encoded OXA-30 β-lactamase. Antimicrob Agents Chemother. 2003;47(7):2380–1.
Sabella C, Tuohy M, Hall G, Gales AC, Erwin ME, Jones RN. Emergence of cefepime-resistance in Klebsiella oxytoca clinical isolate due to alteration in the outer membrane permeability. Clin Microbiol Newsl. 2000;22(5):37–9.
Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466–75.
Okeke I, Laxminarayan R, Bhutta Z, Duse A, Jenkins P, O’Brien T, Pablos-Mendez A, Klugman K. Antimicrobial resistance in developing countries. Part I: recent trends and current status. Lancet Infect Dis. 2005;5(8):481–93.
Nabyonga J, Desmet M, Karamagi H, Kadama P, Omaswa F, Walker O. Abolition of cost-sharing is pro-poor: evidence from Uganda. Health Policy Plan. 2005;20(2):100–8.
Konde-Lule J, Gitta SN, Lindfors A, Okuonzi S, Onama VO, Forsberg BC. Private and public health care in rural areas of Uganda. BMC Int Health Hum Rights. 2010;10:29.
Wang HH, Schaffner DW. Antibiotic resistance: how much do we know and where do we go from here. Appl Environ Microbiol. 2011;77(20):7093–5.
Costelloe C, Metcalfe C, Lovering A, Mant D, Hay AD. Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. BMJ. 2010;340:c2096.
Sethi AK, Acher CW, Kirenga B, Mead S, Donskey CJ, Katamba A. Infection control knowledge, attitudes, and practices among healthcare workers at Mulago Hospital, Kampala, Uganda. Infect Control Hosp Epidemiol. 2012;33(9):917–23.
Allegranzi B, Storr J, Dziekan G, Leotsakos A, Donaldson L, Pittet D. The First Global Patient Safety Challenge “Clean Care is Safer Care”: from launch to current progress and achievements. J Hosp Infect. 2007;65(Suppl 2):115–23.
Naseer U, Haldorsen B, Simonsen GS, Sundsfjord A. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect. 2010;16(2):171–8.
Pitout JD. Enterobacteriaceae that produce extended-spectrum beta-lactamases and AmpC beta-lactamases in the community: the tip of the iceberg? Curr Pharm Des. 2013;19(2):257–63.
Shakya P, Barrett P, Diwan V, Marothi Y, Shah H, Chhari N, Tamhankar A, Pathak A, Lundborg C. Antibiotic resistance among Escherichia coli isolates from stool samples of children aged 3–14 years from Ujjain, India. BMC Infect Dis. 2013;13(1):477.
Bailey JK, Pinyon JL, Anantham S, Hall RM. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol. 2010;59(Pt 11):1331–9.
Young B, Lye D, Krishnan P, Chan S, Leo Y. A prospective observational study of the prevalence and risk factors for colonization by antibiotic resistant bacteria in patients at admission to hospital in Singapore. BMC Infect Dis. 2014;14(1):298.
Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis. 2011;17(7):1216–22.
Goldberg TL, Gillespie TR, Rwego IB, Wheeler E, Estoff EL, Chapman CA. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol Conserv. 2007;135(4):511–7.
Machado E, Coque TM, Cantón R, Sousa João C, Luísa P. Commensal Enterobacteriaceae as reservoirs of extended-spectrumbeta-lactamases, integrons, and sulgenes in Portugal. Front Microbiol. 2013;4:80.
Albrechtova K, Dolejska M, Cizek A, Tausova D, Klimes J, Bebora L, Literak I. Dogs of nomadic pastoralists in Northern Kenya are reservoirs of plasmid-mediated cephalosporin- and quinolone-resistant Escherichia coli, including pandemic clone B2-O25-ST131. Antimicrob Agents Chemother. 2012;56(7):4013–7.
Mesa RJ, Blanc V, Blanch AR, Cortés P, González JJ, Lavilla S, Miró E, Muniesa M, Saco M, Tórtola MT, et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage). J Antimicrob Chemother. 2006;58(1):211–5.
Chung A, Perera R, Brueggemann AB, Elamin AE, Harnden A, Mayon-White R, Smith S, Crook DW, Mant D. Effect of antibiotic prescribing on antibiotic resistance in individual children in primary care: prospective cohort study. BMJ. 2007;335(7617):429.
Dumba R, Kaddu JB, Wabwire Mangen F. Intestinal helminths in Luweero district, Uganda. Afr Health Sci. 2008;8(2):90–6.
Oyoo R, Leemans R, Mol APJ. Future projections of urban waste flows and their impacts in African metropolises cities. Int J Environ Res. 2011;5(3):705–24.
Kwiringira J, Atekyereza P, Niwagaba C, Gunther I. Descending the sanitation ladder in urban Uganda: evidence from Kampala Slums. BMC Public Health. 2014;14:624.
Muyanja C, Nayiga L, Brenda N, Nasinyama G. Practices, knowledge and risk factors of street food vendors in Uganda. Food Control. 2011;22(10):1551–8.
Gedik H, Voss TA, Voss A. Money and transmission of bacteria. Antimicrob Resist Infect Control. 2013;2(1):22.
Blomberg B, Manji K, Urassa W, Tamim B, Mwakagile D, Jureen R, Msangi V, Tellevik M, Holberg-Petersen M, Harthug S, et al. Antimicrobial resistance predicts death in Tanzanian children with bloodstream infections: a prospective cohort study. BMC Infect Dis. 2007;7(1):43.
Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001;32(8):1162–71.
Zaidi AK, Huskins WC, Thaver D, Bhutta ZA, Abbas Z, Goldmann DA. Hospital-acquired neonatal infections in developing countries. Lancet. 2005;365(9465):1175–88.
Kariuki S, Dougan G. Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann NY Acad Sci. 2014;1323:43–55.
Authors’ contributions
CFN conceived of the study, carried out the experiments and drafted the manuscript. MLJ, HK and SYE supervised the study and participated in its design and performed the statistical analysis. DPK helped in drafting of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Hannington Baluku for technical assistance and the participants and health care workers in three study districts. We thank Pragashinee Pillay of the National Health Services Westville Durban, for helping with parallel screen of ESBL-producing enterobacteria by Vitek card.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The study protocol and consent procedure were reviewed and approved by the Research Ethics committee and the Higher Degrees committee of Makerere University Medical School (IRB #-2006-009) and the Uganda National Council for Science and Technology (HS246). All adult participants gave written informed consent before participation. Written informed consent was obtained from parents or guardians of participants less than 18 years. The consent process included storage and use of the collected stool and urine samples for further studies. We obtained assent from participants below the age of 18 years.
Funding
The E-test strips were provided by AB Biodisc Solna, Sweden at discount price. This work was supported in part with funds from the Swedish International Development Agency (SIDA).
Author information
Authors and Affiliations
Corresponding author
Additional files
13104_2016_2049_MOESM2_ESM.pdf
Additional file 2: Table S1: This additional table shows numbers of outcomes of events in relation to the exposure(s). It complements number 15 of the STROBE guideline checklist.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Najjuka, C.F., Kateete, D.P., Kajumbula, H.M. et al. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes 9, 235 (2016). https://doi.org/10.1186/s13104-016-2049-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-016-2049-8