Abstract
Background
To explore and utilize abundant soil microbes and their beneficial functions, the bacterial and fungal compositions in rhizospheres between red- and yellow-fruited tomato varieties were analyzed using high-throughput sequencing technique.
Result
Our results indicated that different soil microbes in rhizospheres of tomatoes were exactly recruited by different color fruit tomatoes. For the reasons as not only soil bacterial community, but also soil fungal compositions were all different between red and yellow fruit tomatoes. For example, Nocardioides, norank_f_norank_o_Vicinamibacterales, norank_f_norank_o_norank_c_KD4-96, norank_f_Birii41, norank_f_norank_o_S085 and Bradyrhizobium were the specific dominant soil bacterial genera, and Lecythophora, Derxomyces and unclassified_f_Pyronemataceae were the dominant soil fungal genera in the rhizospheres of red tomato varieties. By contrast, unclassified_f__Micromonsporaceae, Acidipila, Roseisolibacter, Gaiella and norank_f_Xanthobacteraceae were the unique dominant soil bacterial genera in the rhizospheres of yellow tomato varieties. And unclassified_o__Onygenales, Trichocladium, unclassified_c__Sordariomycetes, Pseudogymnoascus, Acremonium, Oidiodendron, Phialemonium, Penicillium, Phialosimplex were the unique dominant soil fungal genera in rhizospheres of yellow tomato varieties. Moreover, a higher abundance of specific soil bacterial and fungal genera in the rhizosphere was found in rhizospheres of the yellow than those of the red tomato varieties.
Conclusion
Soil bacterial and fungal compositions in rhizospheres between red- and yellow-fruited tomato varieties were found significantly different which growing in the same environment under the identical managements. It suggested that different soil microbes in rhizospheres exactly were recruited by different phenotypes tomato varieties related to fruit color formation.
Similar content being viewed by others
Background
Tomato (Lycopersicon esculentum Mill.) fruits may have a wide range of colors, such as red, yellow, purple and orange [1]. As is well known, fruit color is one of the most important commercial qualities of tomatoes because it is used as one of the important factors in the evaluation of the nutritional quality of tomatoes [2].
The range of tomato fruit colors depends mainly on the composition and proportion of pigments contained in the fruit. Carotenoids and anthocyanins are the main pigments in tomato fruit [3]. Previous studies have shown that abscisic acid promotes carotenoid biosynthesis in tomatoes [4]; ethylene also showed a function in promoting carotenoid accumulation in tomatoes [5]. Moreover, auxin could also promote lycopene synthesis and enhance red colors in tomato fruits [6]. Moreover, previous studies have shown that erythromycin influences the synthesis of anthocyanins [7]. Strigolactone has a positive effect on anthocyanin accumulation [8], and jasmonic acid and abscisic acid can synergize with sugars in the anthocyanin synthesis pathway [9].
It is well known that microbes play an important role in soil, particularly in the rhizospheres of plants, and have interdependent relationships with plants [10, 11]. Among them, soil bacteria can produce many plant growth regulators [12, 13], such as auxin [14], gibberellin [15], cytokinin [16], ethylene [17] and abscisic acid [18]. All of the above phytohormones, which are derived from soil microbes, can be taken up by plants and affect plant growth either directly or indirectly by influencing the rhizosphere environment [19, 20]. In addition, fungi also closely interact with plants, producing various phytohormones affecting the endogenous hormone levels of the plants [21], such as auxin [22], abscisic acid [23], jasmonic acid and salicylic acid [24], ethylene [25, 26], erythromycin [27], and cytokinin [28].
Although fruit color formation has been linked to microbes, the kinds of microbes in soil that are related to carotenoid or anthocyanin biosynthesis in tomato fruits are still unclear. Therefore, to elucidate what kinds of soil microbes are related to carotenoid or anthocyanin biosynthesis, the compositions of the soil microbial community in the rhizospheres of yellow and red fruit tomatoes were analyzed.
Results
Soil bacterial diversity and richness in rhizospheres of tomato varieties with different fruit colors
The Ace and Chao1 indices, which describe soil bacterial richness, were not significant differences in rhizospheres of yellow and red tomato varieties and background. Moreover, the soil bacterial diversity, which describes with the Shannon and Simpson indices, were not significant differences among the background, yellow and red tomato varieties too (Table 1).
Compositions of soil bacterial communities in rhizospheres of tomato varieties with different fruit colors
As shown in Fig. 1a, the numbers of dominant soil bacterial phyla (i.e., relative abundances were greater than 1%) among the background, yellow and red tomato varieties were 11, 10, 12, respectively.
Firstly, Proteobacteria (33.43%), Actinobacteriota (18.63%), Firmicutes (17.85%), Chloroflexi (8.73%), Acidobacteriota (8.47%), Bacteroidota (3.38%), Nitrospirae (2.89%), Planctomycetota (1.68%), Myxococcota (1.55%), Patescibacteria (1.47%), Verrucomicrobia (1.29%) and others (1.87%) were the dominant soil bacterial phyla of CK.
In addition, Proteobacteria (30.39%), Actinobacteriota (24.27%), Firmicutes (16.31%), Acidobacteriota (9.40%), Chloroflexi (7.84%), Gemmatimonadota (3.74%), Bacteroidota (2.34%), Myxococcota (1.38%), Planctomycetota (1.32%) and others (1.30%) were the dominant soil bacterial phyla in the rhizosphere of yellow tomato varieties.
In contrast, Proteobacteria (32.59%), Actinobacteriota (19.20%), Firmicutes (15.74%), Chloroflexi (9.83%), Acidobacteriota (9.04%), Gemmatimonadota (2.78%), Bacteroidota (2.44%), Patescibacteria (2.24%), Myxococcota (2.03%), and others (1.64%) were the dominant soil bacterial phyla in the rhizospheres of red tomato varieties.
Among them, Gemmatimonadota, accumulated as the dominant soil bacterial phyla in the rhizospheres of the yellow or red tomato varieties which compared with CK. Patescibacteria and Verrucomicrobia were the unique dominant soil bacterial phyla in the rhizospheres of red tomato varieties. Although the soil bacterial compositions at phylum level in the rhizospheres of the yellow or red tomato varieties were similar to those of CK, but their proportions were different. The result suggested that the soil bacterial compositions at phylum level in rhizospheres of tomatoes strongly followed the soil bacterial compositions in background (CK), but also it indicated that the proportions of soil dominant bacteria in rhizospheres of tomatoes at phylum level could be altered by planting with different tomato varieties. Furthermore, dominant soil bacterial genera (i.e., those with relative abundances were greater than 1%) among the CK, yellow- (Y) and red-fruited tomato varieties (R) numbered 24, 26 and 27, respectively (Fig. 1b).
First, Bacillus (6.57%), Chujaibacter (4.38%), Luteibacter (3.73%), norank_f_norank_o_Gaiellales (3.15%), unclassified_f__Bacillaceae (2.59%), unclassified_f__Xanthobacteraceae (2.38%), norank_f_norank_o_Elsterales (1.98%), norank_o_Vicinamibacterae (1.85%), norank_f_JG30-KF-AS9 (1.81%), Planifilum (1.59%), Mycobacterium (1.47%), norank_f_norank_o_KF-JG30-C25 (1.39%), norank_f_Gemmatiaceae (1.38%), norank_f_norank_o_Vicinamibacterales (1.37%), norank_f_norank_o_norank_c_KD4-96 (1.37%), norank_f_norank_o_Vicinamibacteraceae (1.37%), unclassified_f__Acetobacteraceae (1.37%), Acidibacter (1.35%), norank_f_norank_o_IMCC26256 (1.33%), Streptomyces (1.31%), norank_f_Micropepsaceae (1.23%), Nocardioides (1.04%), Gaiella (1.02%), 1.02 and others (45.05%) were the dominant bacterial genera in CK.
Second, Bacillus (6.02%), unclassified_f__Micromonsporaceae (3.87%), unclassified_f__Xanthobacteraceae (3.79%), Chujaibacter (3.03%), Streptomyces (2.80%), norank_f_67-14 (2.71%), norank_f_norank_o_Gaiellales (2.49%), Acidipila (2.24%), unclassified_f__Bacillaceae (2.02%), norank_f_norank_o_Elsterales (1.90%), norank_f_Micropepsaceae (1.90%), Acidothermus (1.82%), norank_f_JG30-KF-AS9 (1.80%), Planifilum (1.75%), Roseisolibacter (1.60%), Gaiella (1.46%), norank_f_Xanthobacteraceae (1.30%), Hyphomicrobium (1.28%), norank_f_norank_o_IMCC26256 (1.25%), norank_f_Gemmatimonadaceae (1.23%), unclassified_f__Acetobacteraceae (1.20%), Acidibacter (1.15%), Bryobacter (1.11%), norank_f_norank_o_Acidobacteriales (1.08%), norank_f_norank_o_Subgroup_2 (1.02%) and others (42.02%) were the dominant soil bacterial genera in the rhizospheres of yellow tomato varieties.
In contrast, Bacillus (6.32%), norank_f_norank_o_Gaiellales (4.01%), unclassified_f__Xanthobacteraceae (3.67%), Chujaibacter (2.93%), norank_f_JG30-KF-AS9 (2.72%), norank_f_norank_o_Elsterales (2.35%), norank_f_Micropepsaceae (2.13%), Streptomyces (1.90%), unclassified_f__Bacillaceae (1.63%), Planifilum (1.52%), norank_f_67-14 (1.47%), Acidibacter (1.43%), norank_f_norank_o_Subgroup_2 (1.38%), norank_f_norank_o_IMCC26256 (1.33%), Nocardioides (1.30%), norank_f_norank_o_Vicinamibacterales (1.22%), norank_f_Gemmatimonadaceae (1.19%), unclassified_f__Acetobacteraceae (1.17%), norank_f_norank_o_norank_c_KD4-96 (1.16%), Bryobacter (1.13%), Acidothermus (1.07%), Hyphomicrobium (1.07%), norank_f_norank_o_Acidobacteriales (1.04%), norank_f_Birii41 (1.04%), norank_f_norank_o_S085 (1.03%), Bradyrhizobium (1.01%) and others (44.62%) were the dominant soil bacterial genera in the rhizospheres of red tomato varieties.
Based on the above results, Nocardioides, norank_f_norank_o_Vicinamibacterales, norank_f_norank_o_norank_c_KD4-96, norank_f_Birii41, norank_f_norank_o_S085 and Bradyrhizobium were found as the special dominant soil bacterial genera in the rhizospheres of red tomato varieties; By contrast, unclassified_f__Micromonsporaceae, Acidipila, Roseisolibacter, Gaiella and norank_f_Xanthobacteraceae were the unique dominant soil bacterial genera in the rhizospheres of yellow tomato varieties.
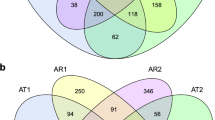
Furthermore, the numbers of soil bacteria obtained in the rhizospheres of yellow and red tomato varieties and CK at the genus level were 832, 836, and 706, respectively (Fig. 2a). Moreover, the numbers of unique bacteria at genus level in rhizospheres of yellow and red tomato varieties and CK were 44, 43 and 7, respectively. In addition, the numbers of soil bacteria obtained at the OTU level in rhizospheres of yellow and red tomato varieties and CK were 4,142, 4,385, and 3,053, respectively. Among them, the numbers of unique bacteria at OTU level in rhizospheres of yellow and red tomato varieties and CK were 301, 420 and 83, respectively (Fig. 2b). These results suggested that the soil bacterial community structure in rhizospheres could be significantly shaped by the tomato varieties, and the numbers of bacteria in the rhizosphere of red fruit tomato varieties were higher than those of yellow fruit tomato varieties. It also indicated that the red fruit tomato varieties recruited more complicated bacteria to help them in accomplishing their growth.
Furthermore, based on the relative abundance data, significant differences in the 15 top soil bacteria at the phylum level in the rhizospheres of different tomato varieties were analyzed using the Wilcoxon rank-sum test. As shown in Fig. 3, they were no significantly different between yellow tomato varieties and CK (Fig. 3a). However, Gemmatimonadota were significantly different between red tomato varieties and CK (Fig. 3b). Furthermore, Actinobacteriota, Patescibacteria and Planctomycetota were significantly different in rhizospheres between yellow and red tomato varieties (Fig. 3c) (Wilcoxon rank-sum test, p < 0.05, p < 0.01).
Moreover, significant differences in the 15 top bacteria at the genus level in the rhizospheres of two different tomato varieties were also analyzed. As shown in Fig. 4, norank_f__67-14 were significantly different between yellow tomato varieties and CK (Fig. 4a). However, they were no significantly different between red tomato varieties and CK (Fig. 4b). But, norank_f__norank_o__Gaiellales, unclassified_f__Micromonosporaceae and norank_f__67-14 were significantly different in the rhizospheres between yellow and red tomato varieties (Fig. 4c) (Wilcoxon rank-sum test, p < 0.05, p < 0.01).
A nonparametric factorial Kruskal–Wallis (KW) rank sum test and LEfSe analysis (LDA threshold of 3.5) were carried out to analyze the significant differences and the main contributing biomarker classes between yellow and red tomato varieties. A greater LDA score indicated a greater influence on species abundance under differential effects (Fig. 5).
As shown in Fig. 5, the soil bacterial compositions were found significantly different in rhizospheres of yellow- and red-fruited tomato varieties along with the CK. Rhodanobacter (genus), Micromonosporales (from order to genus), Solirubrobacterales (from order to genus) were enriched in the rhizospheres of yellow fruit tomato varieties. In contrast, norank_o__Gaiellales (from family to genus), Patescibacteria (phyla), Saccharimonadia (from class to order) were enriched in rhizospheres of red fruit tomato varieties.
In addition, the strongest correlation was detected between the genus norank_f__norank_o__S085 and the other bacteria (Fig. 6). Moreover, most of them were positive connections, such as being positively related to norank_f__norank_o__Vicinamibacterales, norank_f__Gemmatimonadaceae, unclassified_f__Xanthobacteraceae. Hyphomicrobium also showed their own connection networks with other bacteria. For example, norank_f__Gemmatimonadaceae, unclassified_f__Xanthobacteraceae was positively related to norank_f_Chitinophagaceae, but Acidibacter and norank_f__norank_o__Subgroup_2, Bryobacter were more negatively associated with Hyphomicrobium (Fig. 6).
Soil fungal diversity and richness in the rhizospheres of tomato varieties with different fruit color
As seen in Table 2, the Shannon and Simpson indices, which describe the soil fungal diversity in rhizospheres between yellow and red tomato varieties, were not significantly different from each other. Furthermore, the Ace and Chao1 indices, which described the soil fungal richness in the rhizosphere of yellow and red tomato varieties, were also not significantly different from each other (Table 2).
Compositions of soil fungal communities in rhizospheres of tomato varieties with different fruit colors
The numbers of dominant soil fungal phyla (i.e., those with relative abundances greater than 1%) among the background, yellow and red tomato varieties were all 4, respectively.
First, Ascomycota (56.26%), Basidiomycota (35.36%), Mortierellomycota (5.98%), Chytridiomycota and (1.81%) were the dominant soil fungal phyla in CK. Second, Ascomycota (52.04%), Basidiomycota (22.44%), Mortierellomycota (14.46%) and unclassified_k_Fungi (10.78%) were the dominant soil fungal phyla in rhizospheres of yellow tomato varieties. By contrast, Ascomycota (49.63%), Basidiomycota (38.54%), Mortierellomycota (8.26%) and unclassified_k_Fungi (3.36%) were the dominant soil fungal phyla in rhizospheres of red tomato varieties (Fig. 7a).
Furthermore, the d soil ominant fungal genera (i.e., those with relative abundances greater than 1%) among the CK, yellow and red tomato varieties were 11, 19, and 13, respectively.
unclassified_f_Chaetomiaceae (36.26%), unclassified_o__Auriculariales (30.13.%), Mortierella (5.98%), unclassified_c__Agaricomycetes (5.16.%), Penicillium (3.99%), Trichoderma (3.55.%), Sagenomella (3.03%), Aspergillus (2.80%), unclassified_f__Spizellomycetaceae (1.80%), unclassified_o__Sordariales (1.53%), and others (4.00%) were the soil dominant fungal genera in CK.
In addition, unclassified_o__Auriculariales (17.95%), Mortierella (14.46%), unclassified_f__Chaetomiaceae (14.07%), unclassified_k__Fungi (10.78%), Sagenomella (4.97%), unclassified_c__Agaricomycetes (4.09%), unclassified_o__Onygenales (3.46%), Trichocladium (3.41%), Trichoderma (3.21%), unclassified_c__Sordariomycetes (2.70%), Pseudogymnoascus (2.57%), Acremonium (2.41%), Aspergillus (1.98%), Oidiodendron (1.50%), Phialemonium (1.49%), Penicillium (1.32%), unclassified_o__Sordariales (1.21%), Phialosimplex (1.20%), and others (7.06%) were the soil dominant fungal genera in rhizosphere of yellow tomato varieties. By contrast, unclassified_o__Auriculariales (29.12%), unclassified_f__Chaetomiaceae (26.89%), Mortierella (8.25%), unclassified_c__Agaricomycetes (6.67%), Trichoderma (4.14%), unclassified_k__Fungi (3.36%), Lecythophora (2.46%), Sagenomella (2.42%), Derxomyces (2.2%), unclassified_f_Pyronemataceae (1.91%), unclassified_o__Sordariales (1.72%), Aspergillus (1.12%) and others (7.85%) were the soil dominant fungal genera in rhizosphere of red tomato varieties.
Furthermore, Lecythophora, Derxomyces and unclassified_f_Pyronemataceae were the unique soil dominant fungal genera in the rhizospheres of red tomato varieties. However, unclassified_o__Onygenales, Trichocladium, unclassified_c__Sordariomycetes, Pseudogymnoascus, Acremonium, Oidiodendron, Phialemonium, Penicillium, Phialosimplex were the unique soil dominant fungal genera in the rhizosphere of yellow tomato varieties (Fig. 7b).
As seen at Fig. 8, the numbers of soil fungi obtained at the genus level in the rhizospheres of yellow- and red-fruited tomato varieties and CK were 179, 160 and 90, respectively. Moreover, the numbers of unique fungi in the rhizospheres of yellow and red tomato varieties and CK at the genus level were 52, 29 and 2, respectively (Fig. 8a). Similarly, the numbers of soil fungi obtained at the OTU level in the rhizospheres of yellow and red tomato varieties and CK were 540, 516 and 251, respectively. The numbers of unique fungi in the rhizospheres of yellow and red tomato varieties and CK at the OTU level were 163, 134 and 9, respectively (Fig. 8b).
All above results suggested that different soil microorganisms enriched in rhizospheres between red and yellow tomato varieties. In comparison with red tomato varieties, higher abundant of bacteria and fungi were recruited in rhizospheres of yellow tomato varieties.
In addition, significant differences among CK and rhizospheres of yellow and red tomato varieties and the main contributing biomarker classes were also examined by LEfSe analysis (LDA threshold of 3.5).
As shown in Fig. 9, the fungal compositions were significantly different in rhizospheres of yellow and red tomato varieties. Such as Humicola (genus), Trichocladium (genus), Cephalothecaceae (family), Phialemonium (genus), Hypocreales_fam_Incertae_sedis (from family to genus), Cordycipitaceae (from family to genus), Scedosporium (genus), Thelebolales (from order to family), Pseudeurotium (genus), Talaromyces (genus) and unclassified_k__Fungi (from phyla to genus) were enriched in rhizospheres of yellow fruit tomato varieties.
By contrast r, Stemphylium (genus), Pleosporales_fam_Incertae_sedis (family), Microbotryomycetes (genus), Echria (genus) and Sporidiobolales (from class to genus) were significantly enriched in rhizospheres of red tomato varieties. (Fig. 9).
To identify the co-occurrence patterns among species of abundant fungal genera, co-occurrence network analysis was performed.
As shown in Fig. 10, the resulting network showed the strongest correlation between the genus Pseudogymnoascus and the other fungal genera. In addition, Trichocladium had the second strongest correlation to other fungal genera, followed by unclassified_o__Onygenales, Neocosmospora, Wardomyces, Boerlagiomyces, Botryosporium, Cladosporium, Trichoderma and Phialosimplex. They are the top 10 soil fungi with the strongest correlation with other fungi. Most of them correlated positively. Interestingly, the phylum Ascomycota co-occurred with most of the other fungi from genera in the rhizospheres of tomato varieties with different fruit colors.
Genera exhibiting positive or negative correlations can describe the tendency of different fungal genera to co-occur or not in rhizospheres of yellow and red tomato varieties. Therefore, we can speculate that different fungal genera can be recruited by tomato varieties with different fruit colors. Two fungal genera exhibiting a significant positive correlation can grow better through metabolite exchanges, or two genera showing a significant negative correlation in abundance could directly interact through nutrient competition or differ in physiological requirements.
Discussion
The contents of pigments in fruits of different tomato varieties leads to differences in color among their fruits. Studies have shown that the presence of β-carotene, anthocyanins and lycopene in different ratios leads to different colors on the fruit surface. Red tomato fruits have higher contents of lycopene and carotenoid than yellow tomato varieties. In contrast, no lycopene and a higher content of anthocyanin could be detected in yellow tomato varieties than in red tomato varieties [29].
Moreover, numerous studies have shown that plant hormones are closely associated with pigment formation. For example, auxin promotes lycopene accumulation [6]; Ethylene not only regulates carotenoid synthesis affecting fruit color changes [30] but can also increase lycopene content [31]. Moreover, abscisic acid not only regulates the carotenoid content [32] but also promotes lycopene synthesis for fruit reddening [33]. Furthermore, gibberellin acid inhibits carotenoid formation by inhibiting fruit reddening [34]. Likewise, the application of methyl jasmonate increased the β-carotene content and decreased the lycopene content in tomatoes, which affects fruit color changes [35].
On the other hand, abscisic acid has been confirmed to be significantly positively correlated with Actinobacteria and significantly negatively correlated with Proteobacteria [36]. Ethylene can be produced from Nocardioides [37], and auxin can be secreted from Streptomyces [38]. Gibberellin can be synthesized from alpha-Proteobacteria and gamma-Proteobacteria [39, 40], Bradyrhizobium [41], Aspergillus [42, 43] and Penicillium [44]. Moreover, pigments can also be produced by bacteria [45], e.g., Bacillus produces carotenoids [46], lycopene and β-carotene [47]; Trichoderma promoted increases in lycopene [48];
Actinobacteriota and Proteobacteria were found as the dominant soil bacterial phyla in rhizospheres of yellow and red tomato varieties. However, lower abundance of Proteobacteria and higher abundance of Actinobacteriota could be detected in the rhizospheres of yellow tomato varieties than those of red tomato varieties. This result suggested that the sources of abscisic acid in yellow tomato varieties could be speculated more abundant than that of red tomato varieties according to its correlation with Proteobacteria and Actinobacteria. Moreover, Nocardioides, which related to ethylene production,were detected as the unique dominant soil bacterial genera, Streptomyces and Bacillus was more abundant in the rhizospheres of red tomato varieties. Furthermore, Trichoderma was more abundant in the rhizospheres of red tomato varieties and showed prominent roles in cooccurrence network analysis. Based on these microbial functions, higher contents of auxin and ethylene in red-fruited tomato varieties than in yellow-fruited varieties could also assumed.
The abundance of Actinobacteriota in rhizosphere of yellow tomato varieties was also higher than that of red tomato varieties and showed a significant contribution to yellow tomato fruit color formation in the difference test. At the same time, the abundance of the dominant soil fungal genus Aspergillus was also higher in yellow than that in red tomato varieties. Moreover, Penicillium was the unique dominant soil fungal genera in yellow tomato varieties.
The results showed that Proteobacteria, Aspergillus and Penicillium may increase the endogenous gibberellin content of yellow-colored tomato varieties, and it can be inferred that the gibberellin content of yellow tomato varieties is higher than that of red tomato varieties. In addition, Bradyrhizobium, Aspergillusand and Penicillium are also considered as the gibberellines sources.
All of the above results confirmed that tomato varieties with different fruit colors recruited different functional soil microbes in the rhizosphere to produce plant hormones or were sources of different plant hormones related to fruit color formation.
Conclusions
Nocardioides, norank_f_norank_o_Vicinamibacterales, norank_f_norank_o_norank_c_KD4-96, norank_f_Birii41, norank_f_norank_o_S085 and Bradyrhizobium were the special soil dominant bacterial genera in the rhizospheres of red-fruited tomato varieties. In contrast, unclassified_f__Micromonsporaceae, Acidipila, Roseisolibacter, Gaiella and norank_f_Xanthobacteraceae were the unique soil dominant bacterial genera in the rhizospheres of yellow-fruited tomato varieties. In addition, unclassified_o__Onygenales, Trichocladium, unclassified_c__Sordariomycetes, Pseudogymnoascus, Acremonium, Oidiodendron, Phialemonium, Penicillium, Phialosimplex were the unique soil dominant fungal genera in rhizosphere of yellow tomato varieties. Lecythophora, Derxomyces and unclassified_f_Pyronemataceae were the unique soil dominant fungal genera in rhizospheres of red tomato varieties. Based on the functions of these special dominant soil bacteria and fungi in rhizospheres of yellow and red tomato varieties, it can be concluded that different soil microbes in rhizospheres are recruited by different tomato phenotypes related to tomato fruit color formation.
Methods
Field site description and experimental designs
Two different colors tomato groups were used in this study, which d included four yellow-fruited tomato varieties (Jinniu 101 (a), Huang Xiaoya (b), Jimei No. 3 (c) and Milk Tomato (d)) and four red-fruited tomato varieties (Ally (e), Fengzhu (f), Taotaro (g) and Millenium (h). All above tomato varieties were purchased from Nong You Seedling Company (Fig. 11). All tomato varieties were identically treated and grew in the experimental station of the College of Agriculture, Guangxi University, Nanning (108°17′E and 22°51′N).
Soil sampling and physicochemical properties
Rhizosphere soil samples were randomly collected by shaking method [49] during the fruit ripening stages. Briefly, three plants of each tomato variety were randomly selected, and then the whole plants including roots were dug out using a sterilized shovel. Meanwhile, soil samples from identical fields without any plant growth were also collected using as the CK.
The soil physical and chemical properties of the trial site were as follows: soil pH 5.68, organic matter content 8.92 g·kg−1, total nitrogen 0.55 g·kg−1, total phosphorus 0.67 g·kg−1, and total potassium 7.51 g·kg−1. The contents of alkaline dissolved nitrogen, available phosphorus and potassium were 15.27 mg·kg−1, 0.67 mg·kg−1, and 82.8 mg·kg−1, respectively.
Test methods
Soil physicochemical properties
Soil pH value was determined with a pH meter (soil water ratio 1:2.5, w/ v); and the soil organic matter (SOM) content was determined by an external heating met hod using potassium dichromate [50]. Soil total nitrogen (TN) content was quantified by the Kjeldahl acid digestion method; soil total phosphorus (TP) content was quantified using the molybdate blue method after acid di gestion [51]. Soil total potassium (TK) was determined by alkali fusion flame spectrophotometry; soil available nitrogen (AN), phosphorus (AP) and potassium (AK) were subjected to the alkali diffusion method, double acid method and flame photometry, respectively [52].
Analysis of soil microbial diversity
Total DNA extraction, PCR amplification and sequence determination of the root samples were performed by Shanghai Majorbio Biopharm Technology Co., Ltd. High-throughput sequencing was performed using the MiSeq platform.
Total DNA extraction was performed according to the instructions of the FastDNA® Spin Kit for Soil (MP Biomedicals, U.S.), and DNA concentration and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, U.S.). PCR amplification was performed on an ABI GeneAmp® 9700 with the specific primers and sequencing types shown in Tables 3 and 4.
Sequencing was performed using Illumina's MiSeqPE250 platform (Shanghai Majorbio Biopharm Technology Co., Ltd.). PCR products from the same sample were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), mixed and detected by recovery using a 2% agarose gel. The recovered products were quantified using a Quantus™ Fluorometer (Promega, USA). Library construction was carried out using the NEXTFLEX® Rapid DNA-Seq Kit.
The PCR amplification of 16S rRNA gene was performed as follows: initial denaturation at 95 ℃ for 3 min, followed by 27 cycles of denaturing at 95 ℃ for 30 s, annealing at 55 ℃ for 30 s and extension at 72 ℃for 45 s, and single extension at 72 ℃ for 10 min, and end at 4 ℃. The PCR mixtures contain 5 × TransStart FastPfu buffer 4 μL, 2.5 mM dNTPs 2 μL, forward primer (5 μM) 0.8 μL, reverse primer (5 μM) 0.8 μL, TransStart FastPfu DNA Polymerase 0.4 μL, template DNA 10 ng, and finally ddH2O up to 20 μL. PCR reactions were performed in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to manufacturer’s instructions and quantified using Quantus™ Fluorometer (Promega, USA).
Illumina MiSeq sequencing: PCR products from the same sample were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), mixed and detected by recovery using a 2% agarose gel. The recovered products were quantified using a Quantus™ Fluorometer (Promega, USA). Library construction was carried out using the NEXTFLEX® Rapid DNA-Seq Kit.
The PCR amplification of 16S rRNA gene was performed as follows: initial denaturation at 95 ℃ for 3 min, followed by 27 cycles of denaturing at 95 ℃ for 30 s, annealing at 55 ℃ for 30 s and extension at 72 ℃for 45 s, and single extension at 72 ℃ for 10 min, and end at 4 ℃. The PCR mixtures contain 5 × TransStart FastPfu buffer 4 μL, 2.5 mM dNTPs 2 μL, forward primer (5 μM) 0.8 μL, reverse primer (5 μM) 0.8 μL, TransStart FastPfu DNA Polymerase 0.4 μL, template DNA 10 ng, and finally ddH2O up to 20 μL. PCR reactions were performed in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to manufacturer’s instructions and quantified using Quantus™ Fluorometer (Promega, USA).
Processing of sequencing data: The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 [53] and merged by FLASH version 1.2.7 [54] with the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score of < 20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded, reads containing ambiguous characters were also discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of overlap region is 0.2. Reads that could not be assembled were discarded; (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, exact barcode matching, 2 nucleotide mismatch in primer matching [55].
Operational taxonomic units (OTUs) with 97% similarity cut off [56, 57] were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 [58] against the 16S rRNA database using confidence threshold of 0.7.
Raw data were uploaded to the NCBI database for comparison. The data of the comparison database are as follows: bacterial for Silva (Release138, http://www.arb-silva.de); fungal for Unite (Release 8.0, http://unite.ut.ee/index.php).
Statistical analyses
The data was statistically analyzed using Excel 2019 and Statistical Product and Service Solutions (SPSS) Statistics 21, And the R language (version 3.3.1) tool was used for Venn statistics and graphing. And R language (version 3.3.1) tool stats package and Python scipy package were used for difference test. Linear discriminant analysis (LDA) was performed using LEfSe (http://huttenhower.sph.harvard.edu/galaxy/root?tool_id=lefse_upload) on samples according to different grouping conditions based on taxonomic composition to identify clusters that had a significant differential impact on sample delineation. The results are shown as the means with their standard deviations (means ± SDs). Online data analysis was performed using the free online cloud platform (http://www.majorbio.com) of the Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
Availability of data and materials
Raw data for bacterial and fungal bacterial sequence were deposited in the NCBI Sequence Read Archive (SRA) database under accession number PRJNA 859,555 and PRJNA 859,291, respectively.
References
Kang SI, Rahim MA, Afrin KS, Jung HJ, Kim HT, Park JI, et al. Expression of anthocyanin biosynthesis-related genes reflects the peel color in purple tomato. Hortic Environ Biotechnol. 2018;59:435–45.
Gonzali S, Perata P. Fruit colour and novel mechanisms of genetic regulation of pigment production in tomato fruits. Horticulturae. 2021;7:259.
Borghesi E, Ferrante A, Gordillo B, Rodríguez-Pulido FJ, Cocetta G, Trivellini A, et al. Comparative physiology during ripening in tomato rich-anthocyanins fruits. Plant Growth Regul. 2016;80:207–14.
Mou W, Li D, Bu J, Jiang Y, Khan ZU, Luo Z, et al. Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PLoS ONE. 2016;11: e0154072.
Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, et al. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol. 2015;15:114.
Dou JL, Lu XQ, Liu WG, Zhao SJ, He N, Zhu HJ, et al. Correlations of endogenous hormones and lycopene accumulation during development of different ploidy watermelons. Acta Horticulturae Sinica. 2015;42:969–78.
Weiss D, van Tunen AJ, Halevy AH, Mol JNM, Gerats AGM. Stamens and gibberellic acid in the regulation of flavonoid gene expression in the corolla of petunia hybrida. Plant Physiol. 1990;94:511–5.
Ito S, Nozoye T, Sasaki E, Imai M, Shiwa Y, Shibata-Hatta M, et al. Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis. PLoS ONE. 2015;10: e0119724.
Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–16.
Asemoloye MD, Jonathan SG, Ahmad R. Synergistic plant-microbes interactions in the rhizosphere: a potential headway for the remediation of hydrocarbon polluted soils. Int J Phytoremediation. 2019;21:71–83.
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21.
Costacurta A, Vanderleyden J. Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol. 1995;21:1–18.
Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42:207–20.
Frankenberger, W., T., Brunner, W. Method of Detection of Auxin-Indole-3-Acetic Acid in Soils by High Performance Liquid Chromatography1. Soil Sci Soc Am J. 1983. https://doi.org/10.2136/sssaj1983.03615995004700020012x.
Tien TM, Gaskins MH, Hubbell DH. Plant growth substances produced by Azospirillum brasilense and their effect on the growth of pearl millet Pennisetum Americanum L. Appl Environ Microbiol. 1979;37:1016–24.
Nieto KF, Frankenberger WT. Biosynthesis of cytokinins in soil. Soil Sci Soc Am J. 1989;53:735–735.
Amara U, Khalid R, Hayat R. Soil bacteria and phytohormones for sustainable crop production. Bacterial metabolites in sustainable agroecosystem. 2015.
Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol. 2010;60:579–98.
Arshad M, Frankenberger WT. Microbial production of plant hormones. Plant Soil. 1991;133:1–8.
Arshad M, Frankenberger WT. Plant growth-regulating substances in the rhizosphere: microbial production and functions. Adv Agron. 1997;62:45–151.
Chanclud E, Morel J. Plant hormones: a fungal point of view. Mol Plant Pathol. 2016;17:1289–97.
Gruen HE. Auxins and fungi. Annu Rev Plant Physiol. 2003;10:405–40.
Tudzynski B, Sharon A. Biosynthesis, biological role and application of fungal phytohormones. 2002. https://doi.org/10.1007/978-3-662-10378-4_9.
Chaves FC, Gianfagna TJ. Necrotrophic phase of Moniliophthora perniciosa causes salicylic acid accumulation in infected stems of cacao. Physiol Mol Plant Pathol. 2006;69:104–8.
Smith AM, Cook RJ. Implications of ethylene production by bacteria for biological balance of soil. Nature. https://doi.org/10.1038/252703b0.
Dasilva EJ, Henriksson E, Henriksson LE. Ethylene production by fungi. Plant Sci Lett. 1974;2:63–6.
Tudzynski B, Kawaide H, Kamiya Y. Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr Genet. 1998;34:234.
Cooper SJ, AMA F. Comparison of cytokinin and cytokinin- O -glucoside cleaving β-glucosidase production in vitro by Venturia inaequalis and other phytopathogenic fungi with differing modes of nutrition in planta. Physiol Mol Plant Pathol. 1998;53:61–72.
Ruan MY, Ye QJ, Zhou GZ, Wang RQ, Yao ZP, Li ZM, et al. Changes in pigmentation of cherry tomato fruits of different colours during ripening. J Zhejiang Agricul Scie. 2013;54(5):526–8.
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Shima Y, Nakamura N, et al. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat Plants. 2017. https://doi.org/10.1038/s41477-017-0041-5.
Bartley GE, Scolnik PA, Giuliano G. Molecular Biology of Carotenoid Biosynthesis in Plants. Annu Rev Plant Biol. 1994;45:287–301.
Sun L, Wang YP, Pei C, Ren J, Ji K, Qian L, et al. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J Exp Bot. 2011;62:5659–69.
Liu L, Wei J, Zhang M, Zhang L, Li C, Wang Q. Ethylene independent induction of lycopene biosynthesis in tomato fruits by jasmonates. J Exp Bot. 2012;63(16):5751–61. https://doi.org/10.1093/jxb/ers224.
Yu Y. Studies on main pigment formation and physiological mechanism in tomato by exogenous GA3 and ABA. Shenyang Agricultural University. 2016. https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CDFDLAST2017&filename=1017017310.nh.
Czapski J, Saniewski M. The Effect of Methyl Jasmonate Vapour on Some Characteristics of Fruit Ripening, Carotenoids and Tomatine Changes in Tomato Lycopersicon esculentum Mill. Acta Agrobot. 1995;48:27–35.
Lu YQ. Effects of microbial community on crop growth and phytohormones. Chinese Academy of Agricultural Sciences. 2019. https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CMFD201902&filename=1019074899.nh.
Mattes TE, Coleman NV, Spain JC, Gossett JM. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch Microbiol. 2005;183:95–106.
Abd-Alla M, El-Sayed ES, Rasmey AH. Indole-3-acetic acid (IAA) production by Streptomyces atrovirens isolated from rhizospheric soil in Egypt. J Biol Earth Sci. 2013;3. https://www.researchgate.net/publication/259226300.
Nett RS, Nguyen H, Nagel R, Marcassa A, Charles TC, Friedberg I, et al. Unraveling a tangled skein: evolutionary analysis of the bacterial gibberellin biosynthetic operon. preprint. Microbiol. 2019.
Nagel R, Bieber JE, Schmidt-Dannert MG, Nett RS, Peters RJ. A Third Class: Functional Gibberellin Biosynthetic Operon in Beta-Proteobacteria. Front Microbiol. 2018;9:916.
Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, et al. Gibberellin biosynthesis in bacteria. 2009.
Khan AL, Hamayun M, Kim YH, et al. Gibberellins producing endophytic Aspergillus fumigatus sp. LH02 influenced endogenous phytohormonal levels, isoflavonoids production and plant growth in salinity stress. Process Bioche. 2011;46(2):440–7.
Hamayun M, Khan SA, Khan MA, Khan AL, Kang S, Kim S, et al. Gibberellin production by pure cultures of a new strain of Aspergillus fumigatus. World J Microb Biot. 2009;25(10):1785–92.
Teruko N, Kazumi M, Mami S, Kaori T, Tadako M. Effects of Auxin and Gibberellin on conidial germination and elongation of young hyphae in Gibberella fujikuroi and Penicillium notatum. Plant Cell Physiol. 1985;26(7):1433–7.
Mata-Gómez LC, César Montaez Julio, Méndez-Zavala A, Aguilar CN. Biotechnological production of carotenoids by yeasts: an overview. Microb Cell Factories. 2014;13:1–11.
Khaneja R, Perez-Fons L, Fakhry S, Baccigalupi L, Cutting SM. Carotenoids found in Bacillus. J Appl Microbiol. 2009;108:1889–902.
Obata H, Kawahara H, Yamade K, Ito M. Identification of Carotenoids in an Aerobic Sporeformer(Bacillus alvei) and Culture Conditions. Nippon Shokuhin Kogyo Gakkaishi. 2000;47:163–7.
Mnhs A, Mh B, Jua C, Ssc D. Impact of application of Trichoderma and biochar on growth, productivity and nutritional quality of tomato under reduced N-P-K fertilization - sciencedirect. Ann Agric Sci. 2020;65:107–15.
Riley D, Barber SA. Bicarbonate accumulation and pH changes at the soybean (glycine max (L.) Merr.) root-soil interface. Soil Sci Soc Am J. 1969;33:905.
Li Y, Li Z, Arafat Y, Lin W. Studies on fungal communities and functional guilds shift in tea continuous cropping soils by high-throughput sequencing. Ann Microbiol. 2020;7:70.
Yu S, Wu Z, Xu G, Li C, Wu Z, Li Z, et al. Inconsistent patterns of soil fauna biodiversity and soil physicochemical characteristic along an urbanization gradient. Front Ecol Evol. 2022;9.9:824004.
Xu Y, Du A, Wang Z, Zhu W, Li C, Wu L. Effects of different rotation periods of Eucalyptus plantations on soil physiochemical properties, enzyme activities, microbial biomass and microbial community structure and diversity. For Ecol Manag. 2020;456: 117683.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90.
Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63.
Xiao J, Chen SY, Sun Y, Wu SY, Liang WH, Yang SD. Effects of mechanical weeding on soil fertility and microbial community structure in star anise (Illicium verum Hook.f.) plantations. PLoS ONE. 2022;17(4):e0266949.
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8.
Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44:846–9.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Acknowledgements
We thank Shanghai Majorbio Biopharm Technology Co., Ltd. for sequencing. Thanks for the support of the National Experimental Teaching Demonstration Center of Plant Science, Agricultural College, Guangxi University.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31860547) and the Academic Degree and Graduate Education Reform Project of Guangxi (No. JGY2021013).
Author information
Authors and Affiliations
Contributions
Siyu Chen and Shangdong Yang designed the study. Siyu Chen, Yufei Wei, Yan Sun, Huan Li, and Shangdong Yang collected the samples and the data. Siyu Chen analyzed the data and wrote the draft. Shangdong Yang revised the manuscript. All authors reviewed the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The collection of plant material was complied with relevant institutional, national, and international guidelines and legislation, and permission was obtained from the cultivator.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, S., Sun, Y., Wei, Y. et al. Different rhizosphere soil microbes are recruited by tomatoes with different fruit color phenotypes. BMC Microbiol 22, 210 (2022). https://doi.org/10.1186/s12866-022-02620-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-022-02620-z