Abstract
Background
NIN-like protein (NLP) transcription factors (TFs) compose a plant-specific gene family whose members play vital roles in plant physiological processes, especially in the regulation of plant growth and the response to nitrate-nitrogen. However, no systematic identification or analysis of the NLP gene family has been reported in alfalfa. The recently completed whole-genome sequence of alfalfa has allowed us to investigate genome-wide characteristics and expression profiles.
Results
53 MsNLP genes were identified from alfalfa and renamed according to their respective chromosome distributions. Phylogenetic analysis demonstrated that these MsNLPs can be classified into three groups on the basis of their conserved domains. Gene structure and protein motif analyses showed that closely clustered MsNLP genes were relatively conserved within each subgroup. Synteny analysis revealed four fragment duplication events of MsNLPs in alfalfa. The ratios of nonsynonymous (Ka) and synonymous (Ks) substitution rates of gene pairs indicated that the MsNLP genes underwent purifying selection during evolution. Examination of the expression patterns of different tissues revealed specific expression patterns of the MsNLP genes in the leaves, indicating that these genes are involved in plant functional development. Prediction of cis-acting regulatory elements and expression profiles further demonstrated that the MsNLP genes might play important roles in the response to abiotic stress and in phytohormone signal transduction processes.
Conclusion
This study represents the first genome-wide characterization of MsNLP in alfalfa. Most MsNLPs are expressed mainly in leaves and respond positively to abiotic stresses and hormonal treatments. These results provide a valuable resource for an improved understanding of the characteristics and biological roles of the MsNLP genes in alfalfa.
Similar content being viewed by others
Introduction
NIN-like proteins (NLPs) have been found in many species, such as those of green algae and slime molds, and are considered to belong to a plant-specific family of transcription factors (TFs) [1,2,3,4]. On the basis of previous researchers’ comparisons of NLP gene family members in different species, NLP TFs contain two typical characteristic domains: a highly conserved RWP-RK domain and a PB1 domain located at the C- terminus [5, 6]. The RWP-PK domain is highly conserved and consists of more than 50 amino acids, including characteristic “RWPXRK” residues. The PB1 domain is also highly conserved and consists of approximately 80 amino acids at the C-terminus of the NLP. The RWP-RK domain functions as a DNA-binding domain and alone can bind to nitrate-responsive cis-acting elements independently of nitrate [7, 8]. The PB1 domain is involved in protein–protein interactions by forming a dimer with other genes containing this domain, although some PB1 domains mediate interactions with proteins lacking the PB1 domain [9]. In addition, a highly conserved cGMP phosphodiesterase (GAF) domain was identified at the N-terminus of some NLPs and may be related to signal transduction or dimerization [5, 6].
The earliest research on NLP TFs can be traced back to the legume model plant species Lotus japonicus Nin (nodule inception). First, Nin was used as a nodule-sensing gene and was identified as affecting the early development of nodules [10]. Subsequently, a large number of Nin homologous Nin-like genes were identified in nonlegume plant species, including Arabidopsis [2], rice [5], wheat [11], maize [3] and Brassica napus [12]. In-depth research on NLP TFs has revealed the various domains, regulatory networks and expression patterns of the members of this family to a certain extent. Many studies have confirmed that NLP TFs play important roles in the response to nitrate-nitrogen and in plant growth and development. For example, AtNLP7 functions in nitrate and nitrogen starvation responses by binding to key nitrogen pathway-related genes, including ANR1, NRT1.1, NRT2, and LBD37/38, thus moderating nitrogen assimilation and metabolism by transcriptionally activating or suppressing the expression of downstream genes [13, 14]. AtNLP8 regulates nitrate-promoted seed germination and directly binds to the promoter of an abscisic acid (ABA) catabolism-related enzyme-encoding gene to reduce ABA levels in a nitrate-dependent manner [15]. The expression of Arabidopsis NLP7 can promote plant growth by increasing the assimilation of nitrogen and carbon under both limited and sufficient nitrogen conditions [16]. NLP TFs are induced in response to abiotic stress and participate in the regulation of plant tolerance to abiotic stress. For instance, AtNLP7 knockout plants were more drought resistant than wild- type plants. Therefore, NLP TFs not only play a very important role in regulating nitrogen absorption and assimilation but are also related to plant drought resistance [17]. In addition, it has been confirmed that NLP TFs regulate nitrate-responsive gene expression not only to promote nitrate uptake but also to modulate nodulation [18, 19].
Alfalfa (Medicago sativa L.) is an economically and ecologically important legume herbaceous species that is versatile, is highly productivity, has high feed value and plays potential roles in soil improvement and soil conservation [20,21,22]. So far the basic knowledge of the NLPs is still limited in alfalfa. Because NLP genes are involved in regulating various important physiological processes, it would be highly important to systematically investigate NLP family members in alfalfa. With the release of the whole-genome sequence of alfalfa, we have an opportunity to systematically investigate the evolutionary traits, organization and expression profiles of the NLP gene family members of alfalfa at the genome-wide level [23]. Our results provide insights into the evolutionary history and potential functional diversity of NLP genes in alfalfa.
Results
Identification of the NLP genes in the alfalfa genome
Searching for the RWP-RK conserved domain (PF02042) via an HMM profile, we identified a total of 53 putative NLP family genes in alfalfa, which we named MsNLP1 to MsNLP53. The gene characteristics as well as the chromosome locations and the protein sequence length, molecular weight (MW), isoelectric point (pI), and subcellular locations were shown in Table 1. The lengths of the 53 identified NLPs ranged from 208 to 1312, their MWs ranged from 24.2 to 143.1 kDa, and their pIs ranged from 4.94 to 8.84. The 53 MsNLP genes were unevenly distributed on chromosomes 1 to 8 (Additional file 1). The largest number of genes (24) was found on chromosome 3, while only 1 MsNLP gene was distributed on chromosome 1. The predicted subcellular localization results showed that most of the MsNLPs were localized in the nucleus, and the others were localized in the mitochondria, chloroplasts and cytoplasm, the findings of which were comparable to those of NLPs from other plant species.
Multiple sequence alignment and phylogenetic analysis of the MsNLP genes
The phylogenetic relationship of MsNLPs was examined via multiple sequence alignment of their RWP-RK and PB1 domains in alfalfa and Arabidopsis proteins. Through a SMART protein domain query, it was found that the alfalfa NLP contains two typical RWP-RK and PB1 domains. The RWP-PK domain is highly conserved and consists of approximately 50 amino acids, and all MsNLPs contain “RWPXRK” characteristic residues (Additional file 2). The PB1 domain at the C-terminus is highly conserved and consists of approximately 80 amino acids. The 23 MsNLPs contained a complete PB1 domain, which was partially absent in other members.
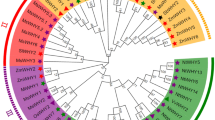
To further explore the evolutionary relationships of the NLP gene family in alfalfa, the sequences of all 9 AtNLPs and 53 MsNLPs were used to construct a phylogenetic tree. Phylogenetic analysis indicated that the alfalfa NLP domains can be divided into three subgroups (named I to III), corresponding to the results of a previous study in Arabidopsis (Fig. 1). Both subclasses I and II contain 8 proteins, with each containing RWP-RK and PB1 domains, which is also the same as AtNLP6/7 and AtNLP8/9 proteins. Subclass III was the largest group, with the 37 remaining members having either both domains or only the RWP-RK domain.
Gene structure and motif composition of the alfalfa NLP gene family
Motif divergence was examined to gain more insight into the evolution of the 53 MsNLPs [24]. A total of 10 motifs were predicted and used to analyze the features of the NLP gene family. The sequence information for each motif is provided in Additional file 3 and Additional file 4. As shown in Fig. 2b, each MsNLP contained between four and ten motifs; some motifs were shared by all members, while others existed in only a few subgroups. For example, motif 1 and motif 8 were present in all members, motif 4 was present in all members except MsNLP17, and motif 7 was detected in only 22 members. Motifs 1 and 8 represent the RWP-RK superfamily domain and RWP-RK domain, respectively. Motif 4 represented the PB1 domain, which was located at the C-terminus (Fig. 2c). These results indicated that the RWP-RK domain and PB1 domain were conserved and specific to plant NLPs. The similar gene structures observed in the same MsNLP subfamily members are consistent with their phylogenetic relationships (Fig. 2a).
The intron diversity divergence of proteins could also provide more insight into the evolution of the NLP gene family in alfalfa. As shown in Fig. 2d, the number of MsNLPs introns ranged from 1 to 10 (2 with one intron, 7 with two introns, 18 with three introns, 14 with four introns, 8 with five introns, 3 with six introns, and 1 with ten introns). The closely related members of each cluster have similar intron structures and small differences in length. Overall, the similarity of conserved motif composition and gene structure of the NLP members in the same groups, combined with the results of phylogenetic analysis, could strongly confirm that the phylogenetic classification was reliable.
Phylogenetic relationships and structure of NLP genes from alfalfa and architecture of their conserved encoded protein motifs. a A phylogenetic tree was constructed by MEGA 6.0 using the neighbor-joining (NJ) method. b Motif composition of NLPs from alfalfa. The motifs, numbered 1–10, are displayed in different colored boxes. c Conserved motif analysis of alfalfa NLP genes. d Exon–intron structure of alfalfa NLP genes. The light green boxes indicate exons, and the black lines indicate introns
Synteny analysis and evolutionary selection pressure of the MsNLP genes
To increase the knowledge of the functions of and mechanisms underlying of NLP gene family members in alfalfa, fragment replication event were investigated. As shown in Fig. 3, four gene pairs (MsNLP7/36, MsNLP8/36, MsNLP15/50 and MsNLP15/53) were distributed on different chromosomes in 69 collinear gene pairs and were considered fragment duplication genes (Additional file 5). These results indicated that MsNLP genes probably originated via gene duplication and that segmental duplication events played a major driving force in MsNLP evolution.
To further analyze the selective constraints among the NLP genes in alfalfa, we calculated the Ka/Ks ratio for the NLP gene pairs. Ka mutations are subject to natural selection, while Ks mutations are not. Neutral selection has occurred when the Ka/Ks ratio equals 1, a Ka/Ks ratio greater than 1 indicates positive selection, while a Ka/Ks ratio less than 1 indicates purifying selection. The distributions of Ka and Ks between each MsNLP pair are shown in Fig. 4. The Ka of MsNLP ranged from 0.001374 to 0.378740, while the Ks ranged from 0.001635 to 1.21454. Among all collinear gene pairs, most pairs for which the Ka/Ks ratio was < 1 were subjected to negative selection, whereas only one was subjected to positive selection (Additional file 6). Taken together, these results suggested that MsNLP genes primarily underwent negative selection; that is, they were subject to purifying selection during evolution.
Cis-acting elements in alfalfa NLP genes
To further explore the potential function of MsNLP genes, the 2000 bp regulatory region upstream of the promoter was selected to analyze the cis-acting elements. Stress-, hormone- and light-responsive elements were detected in the promoter regions of NLP family genes in alfalfa (Fig. 5). Many cis-acting elements related to stress responses, such as LTRs (13), MYBs (254), MYCs (246), MBSs (30), TC-rich repeats (22), and AREs (130), were found in the promoter regions of NLP genes. Hormone-responsive elements such as TGACG motifs (43), TCA elements (54), ABREs (83), P-boxes (49), TGA elements (23), and CGTCA motifs (49) were detected in the promoter regions of MsNLP genes. Moreover, light-responsive elements, for instance, TCCC motifs (18), I-boxes (19), AE-boxes (46), LAMP elements (13), G-boxes (100), ATCT motifs (38), MREs (8), GT1 motifs (110), and Box 4 elements (120), were also found in the promoter of MsNLP genes. Taken together, these results indicated that MsNLP genes may be regulated by cis-acting elements within their promoters during alfalfa growth and in response to stress and phytohormone.
Tissue-specific expression of MsNLPs
To gain a more in-depth understanding of the function of NLP genes in alfalfa development, 10 MsNLPs were randomly selected to analyze their expression in four different tissues (roots, stems, young leaves and mature leaves) via qRT-PCR (Fig. 6). Almost all the genes exhibited tissue-specific transcript accumulation patterns, while the expression profiles of only one gene (MsNLP48) were similar in different tissues. The expression levels of all selected MsNLPs (except MsNLP48) were higher in the leaves (including both young leaves and mature leaves) than in the roots and stems. MsNLP14/24/26/44/47/53 were highly expressed in young leaves, and MsNLP8/30/32 were highly expressed in mature leaves. The MsNLP gene was obviously expressed specifically in the leaves, which indicated that it might be involved in leaf growth and development.
Differential expression of representative MsNLP genes in different tissues, as determined by qRT-PCR. RO: roots; ST: stems; YL: young leaves; MF: mature leaves. The mean expression value was calculated from three independent biological replicates and is expressed in relate to that in the roots. The red represents high expression levels, and the green represents low expression levels
Expression of MsNLPs in response to abiotic stress in leaves
To explore the potential functions of the MsNLP genes in response to different abiotic stresses, the expression profiles of 10 MsNLP genes in response to drought, salt and alkaline stresses were investigated by qRT‒PCR analysis (Fig. 7). The expression levels of some genes tended to constantly fluctuate, which increased in the early stage and then continuously decreased, as was the case for the MsNLP14 gene in response to salt stress. Most genes were expressed at extremely high levels at 12 and 24 h. In general, all MsNLPs were expressed in response to all abiotic treatments. Some genes could be induced by multiple stresses; for example, MsNLP24 was upregulated within 24 h of all tested treatments. Some genes were upregulated until 12 h and then downregulated continuously, as was the case for MsNLP30/47 in response to alkaline stress, whereas other genes were not expressed for 3 h but were upregulated thereafter, as was the case for MsNLP8 in response to drought stress, suggesting that these MsNLP genes might play a crucial role at early and later stages. In addition, some MsNLP genes exhibited opposite expression patterns under different stress conditions. For instance, MsNLP26 was upregulated continuously within 24 h of salt stress treatment but exhibited the opposite expression pattern in response to drought and alkaline stress. In conclusion, MsNLP genes were significantly induced or repressed by the above three treatments.
Expression profiles of 10 selected MsNLP genes in response to different stress treatments. The data were normalized to those of the GAPDH gene. The mean expression values were calculated from three independent biological replicates and are relative to those of the 0 h controls. The different letters indicate that the mean values are significantly different among the treatments (α = 0.05)
MsNLP genes expressed in response to exogenous phytohormones
To confirm whether MsNLP family genes were influenced by different phytohormone treatments, qRT‒PCR experiments were performed to measure the expression levels of MsNLPs in response to treatment with different phytohormones, including abscisic acid (ABA), gibberellin (GA) and indoleacetic acid (IAA) (Fig. 8). Overall, most MsNLP genes were induced by at least one phytohormone treatment; for example, MsNLP 8/24/26/30/44/47/48/53 were induced in response to all tested treatments. In contrast, some MsNLP genes were induced in response to only one or two treatments. For instance, MsNLP14 expression was induced in response to only ABA treatment. Among these MsNLP genes, the expression of some was not significantly changed within 3 h and increased in the later period after phytohormone treatment, as was the case for MsNLP48 and MsNLP53 in response to ABA, so they may be related to later responses to phytohormone treatments; on the other hand, MsNLP48 was upregulated at 3 h and then decreased after IAA treatment, after which its expression was not significantly changed until 24 h, suggesting that this MsNLP gene plays an important role at the early stage of the response to exogenous phytohormones. Interestingly, the expression patterns of the MsNLP genes were opposite in response to phytohormone treatments. For instance, MsNLP47 was significantly induced within 24 h of IAA treatment but was repressed by ABA treatment.
Expression profiles of 10 selected MsNLP genes in response to different phytohormone treatments. The data were normalized to those of the GAPDH gene. The mean expression values were calculated from three independent biological replicates and are relative to those of the 0 h controls. The different letters indicate that the mean values are significantly different among the treatments (α = 0.05)
Discussion
In this study, a total of 53 NLP genes were identified and sequentially named MsNLP1 to MsNLP53 on the basis of their chromosomal location in alfalfa. Previous studies have revealed nine NLP family genes in Arabidopsis, six in rice and nine in maize [3, 5, 11], indicating that different species have different numbers of NLPs. NLP family genes encode large proteins with slightly relatively low pI values in alfalfa (Table 1), consistent with results of other species, which indicates that these TFs are preferentially active under acidic conditions [5, 7, 25]. All MsNLPs identified in this study contain RWP-RK domains; however, only 23 members contain an intact PB1 domain, and the PB1 domain of the rest is partially deleted (Additional file 2), which was caused by sequence fragmentation during evolution or a sequencing error in the alfalfa genome [12]. Therefore, domain gain and loss are divergent forces that drive expansion of the NLP gene family [26, 27].
Phylogenetic analysis indicated that the NLP genes can be divided into three groups in alfalfa (Fig. 1), which is consistent with previous evolutionary classifications in Arabidopsis, rice and maize. The number of MsNLP introns ranged from 1 to 10 in alfalfa, which was consistent with the results found in Arabidopsis, indicating that different species display similarities in terms of the diversity of the structure of the NLP genes [5]. Previous research has shown that segmental duplication can result in a slower rate of intron gain than intron loss [28]. Group II had more introns than group I and group III (Fig. 2), indicating that group II was the most conserved because this group might contain the original genes. Overall, the similarity of conserved motif composition and gene structure of the NLP members in the same groups, combined with the results of phylogenetic analysis, strongly confirm that the phylogenetic classification was reliable. Segmental duplication events are critical for the rapid expansion and evolution of gene families and are essential for environmental adaptability and speciation [29,30,31]. According to our collinearity analysis of the MsNLP genes, we observed all pairs of four groups of fragment duplications (Fig. 3and Additional file 5), in which similar events have been found to have occurred in many species, including Arabidopsis thaliana, rice [5], tomato [32] and others. Therefore, the presence of duplication events implied that segmental duplications were the major evolutionary mechanisms that drove alfalfa NLP expansion in our research. Notably, the Ka/Ks ratio for all the NLP gene pairs except one (MsNLP05/08, which was greater than 1) was less than 1 (Fig. 4and Additional file 6), implicating mainly purifying selection, which favours the elimination of deleterious mutant genes during evolution [33]. This result is consistent with the study of the NLP gene family in maize [3].
Tissue-specific expression patterns of NLP genes in different tissues have been investigated in several plant species. For example, by assessing their expression levels in different tissues of Arabidopsis and rice, researchers have found that NLPs are expressed widely in almost all organs, including seeds, roots, stems, flowers, leaves, nodules, inflorescences, etc [7]. In Arabidopsis, AtNLP8 and AtNLP9 are highly expressed in senescing leaves and seeds compared with other organs, and their expression levels are moderate or very low. OsNLP1 and OsNLP3, whose transcripts constitute the majority of OsNLP transcripts, were found to be preferentially expressed in source organs [5]. It has also been reported that NLPs are highly expressed in the leaves, roots, male catkins, xylem, seeds, and female catkins of Brassica napus [12]. In the present study, the expression levels of most genes except MsNLP48 were higher in the leaves than in the roots and stems. MsNLP14/24/26/44/47/53 were highly expressed in young leaves, whereas MsNLP8/30/32 were highly expressed in mature leaves (Fig. 6), indicating that these genes may participate in leaf growth and development. In addition, we speculate that MsNLPs accumulate in leaves might be used for storing nitrogen to coordinate leaf expansion and photosynthetic capacity to promote leaf growth and biomass, according to previous reports [12, 34]. Moreover, the expression of alfalfa NLPs in the roots was much lower than that in the leaf tissues (in which expression levels were high), indicating that NLPs are involved in mainly nitrate transport rather than the nitrate absorption process [11].
NLPs are essential TFs involved in nitrate signalling [35,36,37]. It has been reported that nitrate triggers nitrate-CPK (Ca2+-sensor protein kinase)-NLP signalling and nitrate-coupled CPK signalling to phosphorylate NLPs, which play a central role in nitrate signalling and integrate transcription, transport, metabolism and systematic growth processes in plants [14, 17, 38]. In our study, analysis of the promoters of the NLP gene family members revealed many cis-acting elements associated with stress- and hormone-responsive elements, suggesting that the MsNLP genes may be regulated by cis-acting elements within their promoters during alfalfa growth and in response to stress and phytohormones (Fig. 5). It has been shown that OsNLP4 is repressed by several abiotic stresses, including drought, cold and submergence in rice [39]. OsNLP3 is induced after germination and repressed by heat treatment [40]. Previous studies have shown that Arabidopsis thaliana lacking AtNLP7 is more drought resistant than the normal wild type, suggesting that NLP expression affects plant tolerance to drought [17]. The expression of SlNLP1, SlNLP2, SlNLP4, and SlNLP6 was upregulated after nitrogen starvation treatment in tomato [41]. OsNLP1, a key gene regulating nitrogen utilization, was rapidly induced by nitrogen starvation in rice [42]. In addition, it was shown that nlp7 mutant plants were hypersensitive to ABA and submergence-induced hypoxia in Arabidopsis thaliana [43]. The expression of LATD/NIP is regulated by hormones, particularly by abscisic acid, which has been previously shown to rescue the primary and lateral root meristem arrest of latd mutants in Medicago truncatula [44]. These results are consistent with the present study, in which MsNLPs were equally responsive to abiotic stresses and hormones in alfalfa. Therefore, the goal of this study was to obtain further insights into the biological functions of the MsNLP genes in response to abiotic stress and phytohormone treatment. Previous research has shown that abiotic stress not only affects alfalfa growth and development but also usually leads to decreases in crop yield and quality [45]. Our results revealed that ten NLP genes were upregulated or downregulated under drought stress, especially at 12 h of prolonged stress (Fig. 7). Combined with the results of previous research that Arabidopsis thaliana AtNLP7 knockout plants were more drought resistant than wild-type plants, the results of the present study indicate that NLP TFs are related to drought resistance in plants [17]. Although MsNLPs exhibited different expression patterns, MsNLPs were expressed in response to both salt and alkaline treatment. In addition, the expression of most MsNLP genes significantly decreased or increased following ABA treatment (Fig. 8). In Arabidopsis, AtNLP8 can directly bind to the promoter of an ABA catabolism-related enzyme-encoding gene to reduce ABA levels, which implies that NLP TFs may participate in the regulation of the ABA signalling pathway. Similarly, some MsNLP genes were induced by GA and IAA phytohormone treatment. Overall, our study lays a foundation for further functional investigation of NLPs in alfalfa.
Conclusions
In summary, 53 NLP genes were identified and further classified into three main groups in alfalfa, with highly conserved intron structures and motif compositions occurring within members of the same group. Synteny analysis and the Ka/Ks ratios revealed that the MsNLP genes underwent fragment duplication events and were subjected to purifying selection during evolution. Tissue expression pattern and expression profile analyses indicated that the MsNLP genes play pivotal roles in alfalfa development and participate in the response to abiotic stress and phytohormone-related signal transduction processes. These results provide a valuable resource for an improved understanding of the characteristics and biological roles of the MsNLP genes in alfalfa.
Materials and methods
Identification and characterization of alfalfa NLP genes
First, the protein sequences of the 9 NLP members of Arabidopsis thaliana were downloaded from a database (http://www.arabidopsis.org) and used as query sequences. Afterward, the published whole-genome protein sequence of cultivated alfalfa (cultivated XinJiangDaYe ) were used to construct a library of the genome database (https://figshare.com/projects/whole_genome_sequencing_and_assembly_of_Medicago_sativa/66380), and the alfalfa genome sequence was then subjected to local BLASTP searches, where the E value was than 1e-5. Subsequently, in accordance with the hidden Markov model (HMM) query strategy, the sequence of the conserved RWP-RK (PF02042) domain of the NLP gene family members was downloaded from the Pfam (http://pfam.xfam.org/) database, after which HMMER 3.0 software was used to search the alfalfa genome and filter genes that were 100% similar and whose E value was less than 1e-5. Finally, after the removal of duplicates and integration of the alfalfa NLP sequence information obtained via the above two procedures, each NLP member was named sequentially according to its distribution on the chromosomes of alfalfa [46]. Physicochemical properties such as the length, molecular weight (MW) and isoelectric point (pI) of the proteins were predicted using the ExPASy ProtParam (www.expasy.org) online analysis tool. In addition, the subcellular locations of alfalfa NLP proteins were predicted by WoLF PSORT (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
Multiple sequence alignment and phylogenetic analysis
The domain sequences of the characterized NLP proteins were used to create multiple protein sequence alignments using ClustalW, with the default parameters [47]. Phylogenetic analysis was performed with MEGA 6.0 using the maximum likelihood method with the following parameters: Poisson model, pairwise deletion, and 1000 bootstrap replications [48].
Gene structure and motif composition of alfalfa NLP gene family members
Conserved domains were analyzed and visualized using CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd) [49, 50]. The conserved motif sequences of the genes were subsequently analyzed by the MEME program (http://meme-suite.org/), and the structure of the MsNLP genes was analyzed with TBtools software [51].
Synteny analysis and evolutionary selection pressure of MsNLP genes
To analyze the syntenic relationship of MsNLP genes, syntenic analysis maps were constructed using the MCScanX software tool. Based on the synteny map of the MsNLP genes, nonsynonymous (Ka) and synonymous (Ks) nucleotide substitutions of each duplicated NLP gene were calculated using KaKs_Calculator 2.0 [52].
Analysis of cis-element within the MsNLP gene promoter
The NLP gene translation start site approximately 2000 bp upstream of ATG from the alfalfa genome was selected as the promoter region, and all the MsNLP promoter sequences were submitted to PlantCARE (http://bioinformatics.psb.ugent.be) to search for plausible cis-acting elements [53].
Plant materials and treatments
Cultivated Medicago sativa L. ‘XinjiangDaYe’, provided by the Xinjiang Academy of Agricultural Sciences, was used in this study. Alfalfa seeds with full grains and consistent morphology were selected for cultivation in vermiculite, and 1/10-strength Hoagland nutrient solution was applied through irrigation during this period. Vigorously growing alfalfa plants were selected for determination of tissue expression specificity and expression patterns in response to various stresses and hormone treatment experiments at 30 d after sowing. The stems, roots, young leaves and mature leaves of alfalfa plants were collected separately for RNA extraction and subjected to quantitative real-time PCR (qRT-PCR) analysis. For drought, salt and alkaline treatments, the plants were subjected to 15% polyethylene glycol (PEG) 6000, 150 mM NaCl and 150 mM NaHCO3 for 3, 6, 12 and 24 h. For phytohormone treatments, 30-d-old seedlings were transplanted into Murashige and Skoog (MS) medium liquid media supplemented with 100 µM ABA, 100 µM GA and 100 µM IAA for 1, 3, 6, 12 and 24 h. Three independent replicates were included for each treatment, and the tissues were collected, immediately frozen in liquid nitrogen and stored at − 80 °C for subsequent analysis.
RNA extraction and gene expression analysis
An Ultrapure RNA Kit (CoWin Biotech, Beijing, China) was used to extract the total RNA from the samples according to the protocol provided by the manufacturer. In accordance with the HiScript II Q Select Reverse Transcriptase Kit (Vazyme Biotech, Nanjing, China) instructions, oligo (dT) 10 reverse primers were used for first-strand cDNA synthesis. Quantitative RT-PCR was carried out according to the method provided by ChamQ™ Universal SYBR qPCR Master Mix (Vazyme, China). The alfalfa GAPDH gene was used as an internal control, and all the primers used are listed in Additional file 7. The reaction was carried out as follows: 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 34 s; and 95 °C for 15 s [54, 55].
Statistical analyses
Each experiment included three independent biological replicates and technical replicates, and all the data are expressed as the means ± standard deviations (SDs). GraphPad Prism 5.0 was used to map the final data, and differences among groups were tested using one-way ANOVA with SPSS 25.0 software. Relative gene expression levels were evaluated according to the 2 −ΔΔCT method.
Data Availability
The data does not involve sequencing and are listed in the article and its additional files.
Abbreviations
- ABA:
-
Abscisic acid
- GA:
-
Gibberellin
- GAF:
-
cGMP phosphodiesterase
- HMM:
-
Hidden Markov model
- IAA:
-
Indoleacetic acid
- Ka:
-
Nonsynonymous substitution ratio
- Ks:
-
Synonymous substitution ratio
- MS:
-
Murashige and Skoog
- MW:
-
Molecular weight
- NJ:
-
Neighbor-joining
- NLP:
-
NIN-LIKE PROTEIN
- PEG:
-
Polyethylene glycol
- pI:
-
Isoelectric point
- qRT-PCR:
-
Quantitative real-time PCR
- TFs:
-
Transcription factors
References
Koi S, Hisanaga T, Sato K, Shimamura M, Yamato KT, Ishizaki K, Kohchi T, Nakajima K. An evolutionarily conserved plant RKD factor controls germ cell differentiation. Curr Biol. 2016;26(13):1775–81.
Mu X, Luo J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell Mol Life Sci. 2019;76(19):3753–64.
Ge M, Liu Y, Jiang L, Wang Y, Lv Y, Zhou L, Liang S, Bao H, Zhao H. Genome-wide analysis of maize NLP transcription factor family revealed the roles in nitrogen response. Plant Growth Regul. 2017;84(1):95–105.
Wang Z, Zhang L, Sun C, Gu R, Mi G, Yuan L. Phylogenetic, expression and functional characterizations of the maize NLP transcription factor family reveal a role in nitrate assimilation and signaling. Physiol Plant 2018.
Schauser L, Wieloch W, Stougaard J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol. 2005;60(2):229–37.
Konishi M, Yanagisawa S. Emergence of a new step towards understanding the molecular mechanisms underlying nitrate-regulated gene expression. J Exp Bot. 2014;65(19):5589–600.
Chardin C, Girin T, Roudier F, Meyer C, Krapp A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J Exp Bot. 2014;65(19):5577–87.
Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617.
Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007(401):re6.
Xiao A, Yu H, Fan Y, Kang H, Ren Y, Huang X, Gao X, Wang C, Zhang Z, Zhu H, et al. Transcriptional regulation of NIN expression by IPN2 is required for root nodule symbiosis in Lotus japonicus. New Phytol. 2020;227(2):513–28.
Kumar A, Batra R, Gahlaut V, Gautam T, Kumar S, Sharma M, Tyagi S, Singh KP, Balyan HS, Pandey R, et al. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L). PLoS ONE. 2018;13(12):e0208409.
Liu M, Chang W, Fan Y, Sun W, Qu C, Zhang K, Liu L, Xu X, Tang Z, Li J et al. Genome-wide identification and characterization of NODULE-INCEPTION-Like protein (NLP) family genes in Brassica napus. Int J Mol Sci 2018, 19(8).
Jian W, Zhang D-w, Zhu F, Wang S-x, Zhu T, Pu X-j, Zheng T, Feng H, Lin H. -h: nitrate reductase-dependent nitric oxide production is required for regulation alternative oxidase pathway involved in the resistance to Cucumber mosaic virus infection in Arabidopsis. Plant Growth Regul. 2015;77(1):99–107.
Marchive C, Roudier F, Castaings L, Brehaut V, Blondet E, Colot V, Meyer C, Krapp A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713.
Yan D, Easwaran V, Chau V, Okamoto M, Ierullo M, Kimura M, Endo A, Yano R, Pasha A, Gong Y, et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat Commun. 2016;7:13179.
Yu LH, Wu J, Tang H, Yuan Y, Wang SM, Wang YP, Zhu QS, Li SG, Xiang CB. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci Rep. 2016;6:27795.
Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57(3):426–35.
Nishida H, Tanaka S, Handa Y, Ito M, Sakamoto Y, Matsunaga S, Betsuyaku S, Miura K, Soyano T, Kawaguchi M et al. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat Commun 2018, 9(1).
Lin JS, Li X, Luo Z, Mysore KS, Wen J, Xie F. NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nat Plants. 2018;4(11):942–52.
Zhang LL, Zhao MG, Tian QY, Zhang WH. Comparative studies on tolerance of Medicago truncatula and Medicago falcata to freezing. Planta. 2011;234(3):445–57.
Gréard C, Barre P, Flajoulot S, Santoni S, Julier B. Sequence diversity of five Medicago sativa genes involved in agronomic traits to set up allele mining in breeding. Mol Breeding 2018, 38(12).
Cui G, Chai H, Yin H, Yang M, Hu G, Guo M, Yi R, Zhang P. Full-length transcriptome sequencing reveals the low-temperature-tolerance mechanism of Medicago falcata roots. BMC Plant Biol. 2019;19(1):575.
Chen H, Zeng Y, Yang Y, Huang L, Tang B, Zhang H, Hao F, Liu W, Li Y, Liu Y, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat Commun. 2020;11(1):2494.
Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20(10):461–4.
Cheng F, Wu J, Wang X. Genome triplication drove the diversification of Brassica plants. Hortic Res. 2014;1:14024.
Kun-Lu, Wu, et al. Z-JG: the WRKY family of transcription factors in Rice and Arabidopsis and their Origins. DNA Res. 2005;12:9–26.
Wu Z, Liu H, Huang W, Yi L, Qin E, Yang T, Wang J, Qin R. Genome-wide identification, characterization, and regulation of RWP-RK Gene Family in the Nitrogen-Fixing clade. Plants (Basel) 2020, 9(9).
Zhang M, Liu Y, He Q, Chai M, Huang Y, Chen F, Wang X, Liu Y, Cai H, Qin Y. Genome-wide investigation of calcium-dependent protein kinase gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2020;21(1):72.
Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1.
Vision TJ, et al. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–7.
Zhang M, Liu Y, Shi H, Guo M, Chai M, He Q, Yan M, Cao D, Zhao L, Cai H, et al. Evolutionary and expression analyses of soybean basic leucine zipper transcription factor family. BMC Genomics. 2018;19(1):159.
Liu M, Zhi X, Wang Y, Wang Y. Genome-wide survey and expression analysis of NIN-like protein (NLP) genes reveals its potential roles in the response to nitrate signaling in tomato. BMC Plant Biol. 2021;21(1):347.
Nekrutenko A, Makova KD, Li WH. The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 2002;12(1):198–202.
Liu T, Ren T, White PJ, Cong R, Lu J. Storage nitrogen co-ordinates leaf expansion and photosynthetic capacity in winter oilseed rape. J Exp Bot. 2018;69(12):2995–3007.
Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136(1):2483–99.
Jagadhesan B, Sathee L, Meena HS, Jha SK, Chinnusamy V, Kumar A, Kumar S. Genome wide analysis of NLP transcription factors reveals their role in nitrogen stress tolerance of rice. Sci Rep. 2020;10(1):9368.
Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci U S A. 2017;114(9):2419–24.
Liu KH, Niu Y, Konishi M, Wu Y, Du H, Sun Chung H, Li L, Boudsocq M, McCormack M, Maekawa S, et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature. 2017;545(7654):311–6.
Chardin C, Girin T, Roudier F. The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. Journal of Experimental Botany, 65(19).
Chen Y, Wang J, Nguyen NK, Hwang BK, Jwa NS. The NIN-Like protein OsNLP2 negatively regulates ferroptotic cell death and Immune responses to Magnaporthe oryzae in Rice. Antioxid (Basel), 11(9):1795.
Liu M, Zhi X, Wang Y, Wang Y. Genome-wide survey and expression analysis of NIN-like protein (NLP) genes reveals its potential roles in the response to nitrate signaling in tomato. BMC Plant Biol, 21(1):347.
Alfatih A, Wu J, Zhang ZS, Xia JQ, Jan SU, Yu LH, Xiang CB. Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J Exp Bot, 71(19):6032–42.
Castillo MC, Costa-Broseta Á, Gayubas B, León J. NIN-like protein7 and PROTEOLYSIS6 functional interaction enhances tolerance to sucrose, ABA, and submergence. Plant Physiol, 187(4):2731–48.
Yendrek CR, Lee YC, Morris V, Liang Y, Pislariu CI, Burkart G, Meckfessel MH, Salehin M, Kessler H. A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J, 62(1):100–12.
Gou J, Debnath S, Sun L, Flanagan A, Tang Y, Jiang Q, Wen J, Wang ZY. From model to crop: functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotechnol J. 2018;16(4):951–62.
Leng X, Wei H, Xu X, Ghuge SA, Jia D, Liu G, Wang Y, Yuan Y. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics. 2019;20(1):786.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Yang M, Derbyshire MK, Yamashita RA, Marchler-Bauer A. NCBI’s conserved domain database and tools for protein domain analysis. Curr Protoc Bioinformatics. 2020;69(1):e90.
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–3.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative Toolkit developed for interactive analyses of big Biological Data. Mol Plant. 2020;13(8):1194–202.
Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a Toolkit incorporating Gamma-Series methods and sliding window strategies. Genom Proteom Bioinform. 2010;8(1):77–80.
Magail Lescot PD, et al. Gert Thijs,Kathleen Marchal,Yves Moreau: Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Cai H, Zhao L, Wang L, Zhang M, Su Z, Cheng Y, Zhao H, Qin Y. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol. 2017;214(4):1579–96.
Cai H, Zhang M, Chai M, He Q, Huang X, Zhao L, Qin Y. Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol. 2019;221(1):295–308.
Acknowledgements
This study was financially supported by National Natural Science Foundation of China (31872998).
Funding
This research was supported by National Natural Science Foundation of China (31872998). The funding body had no influence on the experimental design, data analysis and interpretation, or writing of the manuscript.
Author information
Authors and Affiliations
Contributions
Guowen Cui and Jinqiu Yu designed the experiments. All authors participated in the experiments. Jinqiu Yu wrote the first draft of the article and Guowen Cui revised it. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yu, J., Yuan, Y., Dong, L. et al. Genome-wide investigation of NLP gene family members in alfalfa (Medicago sativa L.): evolution and expression profiles during development and stress. BMC Genomics 24, 320 (2023). https://doi.org/10.1186/s12864-023-09418-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09418-x