Abstract
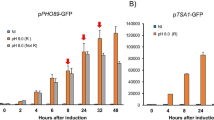
The expression potential of various strains from the Collection of the National Bio-Resource Center (BRC VKPM) belonging to the species Komagataella kurtzmanii, K. phaffii, and K. mondaviorum has been assessed by the level of production of the heterologous enzyme, Citrobacter freundii phytase. Heterologous expression in the K. mondaviorum strains was observed for the first time. We identified the strains of K. phaffii Y-4288, K. mondaviorum Y-4331 and K. phaffii Y-4287 species with a high level production of the heterologous enzyme, a high growth rate, the ability to accumulate a large amount of biomass, and moderate thermotolerance. It has been shown that the average productivity of the transformants based on K. phaffii Y-4288, K. mondaviorum Y-4331, and K. phaffii Y-4287 strains exceeds that of the commercial industrial recipient strain K. phaffii GS115 Y-2837 by more than 3, 5, and 6 times, respectively. The K. phaffii Y-4287 and K. mondaviorum Y-4331 strains exhibited moderate thermotolerance and the ability to accumulate a heterologous product at 37°C. The high expression potential of the identified strains opens up the possibility of creating recipient strains on their basis for high-level production of heterologous proteins.




Similar content being viewed by others
REFERENCES
Rozanov, A.S., Pershina, E.G., Bogacheva, N.V., et al., Diversity and distribution of methylotrophic yeasts used in genetic engineering, Vavilov J. Genet. Breed., 2020, vol. 24, no. 2, pp. 149–157. https://doi.org/10.18699/VJ20.602
Gonçalves, A.M., Pedro, A.Q., Maia, C., et al., Pichia pastoris: a recombinant microfactory for antibodies and human membrane proteins. J. Microbiol. Biotechnol., 2013, vol. 23, no. 5, pp. 587–601. https://doi.org/10.4014/jmb.1210.10063
Schwarzhans, J.P., Wibberg, D., Winkler, A., et al., Integration event induced changes in recombinant protein productivity in Pichia pastoris discovered by whole genome sequencing and derived vector optimization. Microb. Cell Fact., 2016, vol. 15, no. 1, pp. 1–15. https://doi.org/10.1186/s12934-016-0486-7
Yamada, Y., Matsuda, M., Maeda, K., and Mikata, K., The phylogenetic relationships of methanol-assimilating yeasts based on the partial sequences of 18S and 26S ribosomal RNAs: the proposal of Komagataella gen. nov. (Saccharomycetaceae). Biosci. Biotechnol. Biochem., 1995, vol. 59, no. 3, pp. 439–444. https://doi.org/10.1271/bbb.59.439
Dlauchy, D., Tornai-Lehoczki, J., Fulop, L., and Peter, G., Pichia (Komagataella) pseudopastoris sp. nov., a new yeast species from Hungary, Antonie van Leeuwenhoek, 2003, vol. 83, no. 4, pp. 327–332. https://doi.org/10.1023/A:1023318829389
Kurtzman, C.P., Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella, Int. J. Syst. Evol. Microbiol., 2005, vol. 55, no. 2, pp. 973–976. https://doi.org/10.1099/ijs.0.63491-0
Kurtzman, C.P., Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis, J. Ind. Microbiol. Biotechnol., 2009, vol. 36, no. 11, p. 1435. https://doi.org/10.1007/s10295-009-0638-4
Kurtzman, C.P., Komagataella populi sp. nov. and Komagataella ulmi sp. nov., two new methanol assimilating yeasts from exudates of deciduous trees, Antonie van Leeuwenhoek, 2012, vol. 101, no. 4, pp. 859–868. https://doi.org/10.1007/s10482-012-9702-6
Naumov, G.I., Kondratieva, V.I., Meshcheryakova, E.V., and Naumova, E.S., Taxonomic genetics of methylotrophic yeast genus Komagataella: new biological species K. kurtzmanii, Russ. J. Genet., 2016, vol. 52, no. 4, pp. 378–382. https://doi.org/10.1134/S1022795416030108
Naumov, G.I., Naumova, E.S., and Boundy-Mills, K.L., Description of Komagataella mondaviorum sp. nov., a new sibling species of Komagataella (Pichia) pastoris, Antonie Van Leeuwenhoek, 2018, vol. 111, no. 7, pp. 1197–1207. https://doi.org/10.1007/s10482-018-1028-6
Naumov, G.I., Naumova, E.S., Tyurin, O.V., and Kozlov, D.G., Komagataella kurtzmanii sp. nov., a new sibling species of Komagataella (Pichia) pastoris based on multigene sequence analysis, Antonie van Leeuwenhoek, 2013, vol. 104, no. 3, pp. 339–347. https://doi.org/10.1007/s10482-013-9956-7
Gasser, B. and Mattanovich, D., A yeast for all seasons—is Pichia pastoris a suitable chassis organism for future bioproduction?, FEMS Microbiol. Lett., 2018, vol. 365, no. 17, article ID fny181. https://doi.org/10.1093/femsle/fny181
Tyurin, O., Gubaidullin, I., Cheperegin, S., et al., The new expression system based on a novel yeast species of the genus Komagataella, FEBS J., 2013, vol. 280, suppl. S1, article ID SW06, W33-39, p. 602.
Lehnen, M., Ebert, B.E., and Blank, L.M., Elevated temperatures do not trigger a conserved metabolic network response among thermotolerant yeasts, BMC Microbiol., 2019, vol. 19, no. 1, pp. 1–11. https://doi.org/10.1186/s12866-019-1453-3
Manfrão-Netto, J.H.C., Gomes, A.M.V., and Parachin, N.S., Advances in using Hansenula polymorpha as chassis for recombinant protein production, Front. Bioeng. Biotechnol., 2019, vol. 7, p. 94. https://doi.org/10.3389/fbioe.2019.00094
Wallace-Salinas, V. and Gorwa-Grauslund, M.F., Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature, Biotechnol. Biofuels, 2013, vol. 6, no. 1, pp. 1–9. https://doi.org/10.1186/1754-6834-6-151
Gordeeva, T.L., Borshevskaya, L.N., Kalinina, A.N., et al., Comparative analysis in plate test of expression efficiency of genes for bacterial phytases in Pichia pastoris yeast, Biotechnology, 2017, vol. 33, no. 6, pp. 83–88. https://doi.org/10.21519/0234-2758-2017-33-6-83-88
Gordeeva, T.L., Borschevskaya, L.N., and Sineoky, S.P., Improved PCR-based gene synthesis method and its application to the Citrobacter freundii phytase gene codon modification, J. Microbiol. Methods, 2010, vol. 81, no. 2, pp. 147–152. https://doi.org/10.1016/j.mimet.2010.02.013
Sambrook, J., Fritsch, E.F., and Maniatis, T., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, 1989, 2nd ed., pp. 4–1626.
Chen, C.C., Wu, P.H., Huang, C.T., and Cheng, K.J., A Pichia pastoris fermentation strategy for enhancing the heterologous expression of an Escherichia coli phytase, Enzyme. Microb. Technol., 2004, vol. 35, no. 4, pp. 315–320.
Gordeeva, T.L., Borshchevskaya, L.N., Kalinina, A.N., et al., Pichia pastoris yeast transformant producing phytase, RF Patent no. 2701498, 2018.
Tyurin, O.V., Development of a gene expression system based on methylotrophic yeast Komagataella kurtzmanii, Cand. Sci. (Biol.) Dissertation, State Research Institute of Genetics and Selection of Industrial Microorganisms, 2014.
Funding
This work was supported by the Ministry of Science and Higher Education of the Russian Federation (grant no. 075-15-2019-1659 dated October 31, 2019) and was carried out using the resources of the Unique Scientific Facility of the All-Russian Collection of Industrial Microorganisms National Bio-Resource Center, Kurchatov Institute, GOSNIIGENETIKA NRC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals performed by any of the authors.
This article does not contain any studies involving human participants performed by any of the authors outside the scope of people’s normal professional activities.
Additional information
Translated by I. Gordon
Abbreviations: CL, culture liquid; OD600, optical density at a wavelength of 600 nm; PCR, polymerase chain reaction; SDS-PAGE, polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate.
Rights and permissions
About this article
Cite this article
Gordeeva, T.L., Borschevskaya, L.N., Feday, T.D. et al. The Expression Potential of Novel Komagataella Strains. Appl Biochem Microbiol 58, 916–922 (2022). https://doi.org/10.1134/S0003683822080038
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683822080038