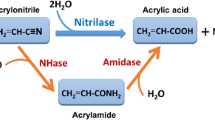
Abstract
A Rhodococcus rhodochrous strain, M33-2nit, has been constructed with two copies of the nitrilase gene from A. denitrificans В-9582 under the control of nitrile hydratase promoter from R. rhodochrous M8. The optimized cultivation of this strain made it possible to obtain the enzyme in a concentration of up to 17 g of dry cells/L with a specific activity of up to 7 U/mg cdw with a two-substrate culturing scheme in a fed-batch reactor with the sequential addition of glucose and acetate. The capacities of A. denitriificans В-9582 and R. rhodochrous M33-2nit cells to synthesize ammonium acrylate from acrylonitrile under conditions imitating industrial synthesis are compared. It is shown that R rhodochrous M33-2nit cells can synthesize ammonium acrylate under higher rates of acrylonitrile feeding than A. denitriificans В-9582 cells. The potential to obtain a highly concentrated solution of ammonium acrylate (450 g/L) with R. rhodochrous M33-2nit cells as a biocatalyst was demonstrated. The conversion of acrylonitrile to ammonium acrylate reached 99.5%.






Similar content being viewed by others
REFERENCES
Bork, P. and Koonin, E.V., A new family of carbon-nitrogen hydrolases, Protein Sci., 1994, vol. 3, no. 8, pp. 1344–1346. https://doi.org/10.1002/pro.5560030821
Pace, H.C. and Brenner, C., The nitrilase superfamily: classification, structure and function, Genome Biol., 2001, vol. 2, no. 1. https://doi.org/10.1186/gb-2001-2-1-reviews0001
Owald, S. and Yanenko, A., Hydrolysis of nitriles to carboxylic acids, in Enzyme Catalysis in Organic Synthesis, 3rd ed., Drauz, K., Groger, H., and May, O., Eds., Weinheim, Germany: Wiley-VCH Verlag GmbH and Co. KGaA, 2012, vol. 2 (3), 14, pp. 545–559. https://doi.org/10.1002/9783527639861.ch14
Alkhazov, T.G., Adzhamov, K.Yu., and Khanmamedova, A.K., Catalytic propylene oxidation, Usp. Khim., 1982, vol. 51, no. 6, pp. 950–967.
Bansal, V., Delgado, Y., Legault, M.D., et al., Low operational stability of enzymes in dry organic solvents: changes in the active site might affect catalysis, Molecules, 2012, vol. 17, no. 2, pp. 1870–1882. https://doi.org/10.3390/molecules17021870
Pirozzi, D. and Halling, P.J., Development of small-size tubular-flow continuous reactors for the analysis of operational stability of enzymes in low-water systems, Biotechnol. Bioeng., 2001, vol. 72, no. 2, pp. 244–248. https://doi.org/10.1002/1097-0290(20000120)72:2<244:: AID-BIT12> 3.0.CO;2-J
Glinskii, S.A., Kozulin, S.V., Kozulina, T.N., et al., Comparative analysis of strains used for obtaining ammonium acrylate, Biotekhnologiya, 2010, no. 1, pp. 17–24.
Novikov, A.D., Ryabchenko, L.E., Leonova, T.E., et al., Bacterial strain Alcaligenes denitrificans C-32 contains two nitrilases with different substrate specificity, Biotekhnologiya, 2016, vol. 32, no.6, pp. 1–8. https://doi.org/10.21519/0234-2758-2016-32-6-45-52
Leonova, T.E., Astaurova, O.B., Ryabchenko, L.E., et al., Nitrile hydratase of Rhodococcus: optimization of synthesis in cells and industrial applications for acrylamide production, Appl. Biochem. Biotechnol., 2000, vol. 88, nos. 1–3, pp. 231–241. https://doi.org/10.1385/ABAB:88:1-3:231
Kobayashi, M., Goda, M., and Shimizu, S., Nitrilase catalyzes amide hydrolysis as well as nitrile hydrolysis, Biochem. Biophys. Res. Commun., 1998, vol. 253, no. 3, pp. 662–666. https://doi.org/10.1006/bbrc.1998.9834
Yamada, H. and Kobayashi, M., Nitrile hydratase and its application to industrial production of acrylamide, Biosci. Biotechnol. Biochem., 1996, vol. 60, no. 9, pp. 1391–1400. https://doi.org/10.1271/bbb.60.1391
Lavrov, K.V., Shemyakina, A.O., Grechishnikova, E.G., et al., New cblA gene participates in regulation of cobalt-dependent transcription of nitrile hydratase genes in Rhodococcus rhodochrous,Res. Microbiol., 2018, vol. 169, nos. 4–5, pp. 227–236. https://doi.org/10.1016/j.resmic.2018.03.006
Simon, R., Priefer, U., and Puhler, A.A., Broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria, Nat. Biotechnol., 1983, vol. 1, no. 9, pp. 784–791. https://doi.org/10.1038/nbt1183-784
Ryabchenko, L.E., Polyakova, I.N., and Yanenko, A.S., Transposable plasmid vectors capable of conjugative transfer between E. coli and Rhodococcus cells and their use for constructing Rhodococcus strains, Biotekhnologiya, 2005, no. 5, pp. 6–13.
Gentz, R., Langner, A., Chang, A.C., et al., Cloning and analysis of strong promoters is made possible by the downstream placement of a RNA termination signal, Proc. Natl. Acad. Sci. U. S. A., 1981, vol. 78, no. 8, pp. 4936–4940.
Tani, K., Kobayashi, T., Sakotani, A., et al., Expression of the gyrB gene as an indicator of growth activity of Escherichia coli,J. Environ. Biotechol., 2012, vol. 12, no. 1, pp. 33–38.
Byrne, G.A., Russell, D.A., Chen, X., et al., Transcriptional regulation of the virR operon of the intracellular pathogen Rhodococcus equi,J. Bacteriol., 2007, vol. 189, no. 14, pp. 5082–5089. https://doi.org/10.1128/jb.00431-07
Livak, K.J. and Schmittgen, T.D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C (T)) Method, Methods, 2001, vol. 25, no. 4, pp. 402–408. https://doi.org/10.1006/meth.2001.1262
Lavrov, K.V., Novikov, A.D., Ryabchenko, L.E., and Yanenko, A.S., Expression of acylamidase gene in Rhodococcus erythropolis strains, Russ. J. Genet., 2014, vol. 50, no. 9, pp. 1003–1008.
Tarutina, M.G., Raevskaya, N.M., Shustikova, T.E., et al., Evaluation of the effectiveness of Corynebacterium glutamicum promoters and their use to enhance gene activity in lysine-producing bacteria, Biotekhnologiya, 2015, no. 6, pp. 16–24.
Yanenko, A.S., Astaurova, O.B., Gerasimova, T.V., et al., Regulation of utilization of nitriles in Rhodococcus,Biotekhnologiya, 1995, no. 7-8, pp. 139–144.
Lorents, I., Voronin, S.P., Kozulin, S.V., et al., RF Patent No. 2340667, 2008.
Shiu, C., Zhang, Z., and Thomas, C.R., A comparison of the mechanical properties of different bacterial species, in Applied Microbiology, Durieux, A. and Simon, J.-P., Eds., Dordrecht, Netherlands: Springer, 2002, vol. 11, pp. 155–162. https://doi.org/10.1007/0-306-46888-3
Keshavarz, E., Hoare, M., and Dunnill, P., Biochemical engineering aspects of cell disruption, in Separations for Biotechnology, Verrall, M.S. and Hudson, M.J., Eds., Chichester, England: Ellis Horwood, 1987, vol. 3, pp. 62–79.
Edebo, L., Disintegration of cells, in Fermentation Advances, Perlman, D., Ed., London, England : Academic, 1969, pp. 249–271.
Sutcliffe, I.C., Brown, A.K., and Dover, L.G., The rhodococcal cell envelope: composition, organisation and biosynthesis, in Biology of Rhodococcus, Alvarez, H.M., Ed., Germany, Berlin: Springer-Verlag, 2010, pp. 29–71. https://doi.org/10.1007/978-3-642-12937-7_2
Funding
This work was supported by the Russian Science Foundation (project no. 16-14-00216, “Study on cobalt-dependent gene expression in Rhodococcus bacteria and the construction on its basis of a platform for the biosynthesis of enzyme biocatalysts to obtain acrylic monomers”).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
SUPPLEMENTARY MATERIALS
The electronic version of the article contains Supplementary Materials, which are available free of charge at the website of the journal Biotechnology, http://www.biotechnology-journal.ru.
Additional information
Translated by I.Gordon
Abbreviations: cdw—cell dry weight; EDTA, ethylene diamine tetraacetic acid; GS, gas chromatography; HPLC, high-performance liquid chromatography; LB medium, lysogeny broth medium; MS medium, mineral synthetic medium; OD600, optical density at wavelength of 600 nm; PAGE, polyacrylamide gel electrophoresis; SD, standard deviation; SDS, sodium dodecyl sulfate.
Rights and permissions
About this article
Cite this article
Lavrov, K.V., Grechishnikova, E.G., Shemyakina, A.O. et al. Optimization of the Expression of Nitrilase from Alcaligenes denitrificans in Rhodococcus rhodochrous to Improve the Efficiency of Biocatalytic Synthesis of Ammonium Acrylate. Appl Biochem Microbiol 55, 861–869 (2019). https://doi.org/10.1134/S0003683819090035
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683819090035