Abstract
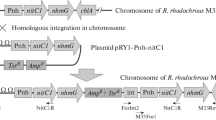
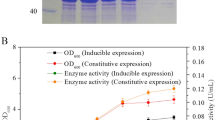
Rhodococcus ruber TH was selected as a parent strain to engineer for biomanufacturing of ammonium acrylate; the characteristics of this strain included accelerated growth rate, high cell tolerance and natively overexpressed nitrile hydratase (NHase). Transcriptome analysis revealed that the transcription levels of the native NHase, amidase and nitrilase were extremely high, moderate and extremely low, respectively. Through NHase-amidase double-knockout and amidase single-knockout, the engineered strains R. ruber THdAdN and R. ruber THdA were obtained for overexpression of a heterologous nitrilase from R. rhodochrous tg1-A6 using a urea-induced Pa2 promoter. The nitrilase activity toward substrate acrylonitrile in the engineered THdAdN(Nit) reached 187.0 U/mL at 42 h, threefold of that R. rhodochrous tg1-A6 and 2.3-fold of that of THdA(Nit). The optimal catalysis temperature and pH of the nitrilases in different cells exhibited no significant difference. Using the cells as catalysts, biomanufacturing of ammonium acrylate was performed under room temperature. When catalyzed by the engineered THdAdN(Nit), the titer and productivity of ammonium acrylate dramatically increased to 741.0 g/L and 344.9 g/L/h, which are the highest results reported to date.






Similar content being viewed by others
References
Agarwal A, Nigam VK (2014) Nitrilase mediated conversion of Indole-3-acetonitrile to Indole-3-acetic acid. Biocatal Agric Biotechnol 3:351–357
Asano Y, Yasuda T, Tani Y, Yamada H (1982) A new enzymatic method of acrylamide production. Agric Biol Chem 46:1183–1189
Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K (2012) Engineering the third wave of biocatalysis. Nature 485:185–194
Chen H, Chen Z, Ni Z, Tian R, Zhang T, Jia J, Chen K, Yang S (2015) Display of Thermotoga maritima MSB8 nitrilase on the spore surface of Bacillus subtilis using out coat protein CotG as the fusion partner. J Mol Catal B Enzym 123:73–80
Chiba K, Hoshino Y, Ishino K, Kogure T, Mikami Y, Uehara Y, Ishikawa J (2007) Construction of a pair of Practical Nocardia-Escherichia coli shuttle vectors. Jpn J Infect Dis 60:45–47
Chu HS, Ahn JH, Yun J, Choi IS, Nam TW, Cho KM (2015) Direct fermentation route for the production of acrylic acid. Metab Eng 32:23–29
Cobb RE, Wang Y, Zhao H (2015) High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. Acs Synth Biol 4:723–728
Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343
Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact 11:1–18
Jiang G, Chen J, Thu HY, Huang JS, Zhu N, Che CM (2009) Microbial transformation of nitriles to high-value acids or amides. Adv Biochem Eng Biotechnol 113:33–77
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239
Kamal A, Kumar MS, Kumar CG, Shaik T (2011) Bioconversion of acrylonitrile to acrylic acid by Rhodococcus ruber strain AKSH-84. J Microbiol Biotechnol 21:37–42
Kim S-H, Oriel P (2000) Cloning and expression of the nitrile hydratase and amidase genes from Bacillus sp. BR449 into Escherichia coli. Enzyme Microb Technol 27:492–501
Komeda H, Kobayashi M, Shimizu S (1996) Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc Natl Acad Sci 93:4267–4272
Leonova TE, Astaurova OB, Ryabchenko LE, Yanenko AS (2000) Nitrile hydratase of Rhodococcus. Appl Biochem Biotechnol 88:231–241
Liu C, Yu H, Ma Y, Pan W, Luo H (2009) Promoter recognition and β-galactosidase reporter gene expression in Rhodococcus. Chin J Biotechnol 25:1360–1365
Luo H, Ma J, Chang Y, Yu H, Shen Z (2015) Directed evolution and mutant characterization of nitrilase from Rhodococcus rhodochrous tg1-A6. Appl Biochem Biotechnol 178(8):1510–1521
Luo H, Lu F, Chang Y, Ma J, Yu H, Shen Z (2008) Gene cloning, overexpression, and characterization of the nitrilase from Rhodococcus rhodochrous tg1-A6 in E. coli. Appl Biochem Biotechnol 160:393–400
Luo H, Wang TG, Hui-Min YU, Yang HF, Shen ZY (2006) Expression and catalyzing process of the nirilase in Rhodococcus rhodochrous tg1-A6. Modern Chem Ind 26:109–113
Ma Y, Yu H (2012) Engineering of Rhodococcus cell catalysts for tolerance improvement by sigma factor mutation and active plasmid partition. J Ind Microbiol Biotechnol 39:1421–1430
Ma Y, Yu H, Pan W, Liu C, Zhang S, Shen Z (2010) Identification of nitrile hydratase-producing Rhodococcus ruber TH and characterization of an amiE-negative mutant. Bioresour Technol 101:285–291
Martínková L, Křen V (2010) Biotransformations with nitrilases. Curr Opin Chem Biol 14:130–137
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177
Nagasawa T, Nakamura T, Yamada H (1990) Production of acrylic acid and methacrylic acid using Rhodococcus rhodochrous J1 nitrilase. Appl Microbiol Biotechnol 34:322–324
Nagasawa T, Shimizu H, Yamada H (1993) The superiority of the third-generation catalyst, Rhodococcus rhodochrous J1 nitrile hydratase, for industrial production of acrylamide. Appl Microbiol Biotechnol 40:189–195
Nagasawa T, Takeuchi K, Nardi-Dei V, Mihara Y, Yamada H (1991) Optimum culture conditions for the production of cobalt-containing nitrile hydratase by Rhodococcus rhodochrous J1. Appl Microbiol Biotechnol 34:783–788
Ni K, Wang H, Zhao L, Zhang M, Zhang S, Ren Y, Wei D (2013) Efficient production of (R)-(−)-mandelic acid in biphasic system by immobilized recombinant E. coli. J Biotechnol 167:433–440
Nicholson EB, Concaugh EA, Mobley HL (1991) Proteus mirabilis urease: use of a ureA-lacZ fusion demonstrates that induction is highly specific for urea. Infect Immun 59:3360–3365
Okamoto S, Petegem FV, Patrauchan MA, Eltis LD (2010) AnhE, a metallochaperone involved in the maturation of a cobalt-dependent nitrile hydratase. J Biol Chem 285:25126–25133
Rustler S, Müller A, Windeisen V, Chmura A, Fernandes B, Kiziak C, Stolz A (2007) Conversion of mandelonitrile and phenylglycinenitrile by recombinant E. coli cells synthesizing a nitrilase from Pseudomonas fluorescens EBC191. Enzyme Microb Technol 40:598–606
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Schmid A, Dordick J, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268
Shi Y, Yu H, Sun X, Tian Z, Shen Z (2004) Cloning of the nitrile hydratase gene from Nocardia sp. in Escherichia coli and Pichia pastoris and its functional expression using site-directed mutagenesis. Enzyme Microb Technol 35:557–562
Sohoni SV, Nelapati D, Sathe S, Javadekar-Subhedar V, Gaikaiwari RP, Wangikar PP (2015) Optimization of high cell density fermentation process for recombinant nitrilase production in E. coli. Bioresour Technol 188:202–208
Sun X, Shi Y, Yu H, Shen Z (2004) Bioconversion of acrylnitrile to acrylamide using hollow-fiber membrane bioreactor system. Biochem Eng J 18(3):239–243
Sun X, Yu H, Shen Z (2009) Deactivation kinetics of nitrile hydratase in free resting cells. Chin J Chem Eng 17(5):822–828
Tan Y, Xu D, Ye L, Wang X (2012) Construction of a novel sacB-based system for marker-free gene deletion in Corynebacterium glutamicum. Plasmid 67:44–52
Vejvoda V, Kubac D, Davidova A, Kaplan O, Sulc M (2010) Purification and characterization of nitrilase from Fusarium solani IMI196840. Process Biochem 45:1115–1120
Wohlgemuth R (2010) Biocatalysis—key to sustainable industrial chemistry. Curr Opin Biotechnol 21:713–724
Wu S, Fallon R, Payne M (1997) Over-production of stereoselective nitrile hydratase from Pseudomonas putida 5B in Escherichia coli: activity requires a novel downstream protein. Appl Microbiol Biotechnol 48:704–708
Wu S, Fogiel AJ, Petrillo KL, Jackson RE, Parker KN, DiCosimo R, Ben-Bassat A, O’Keefe DP, Payne MS (2008) Protein engineering of nitrilase for chemoenzymatic production of glycolic acid. Biotechnol Bioeng 99:717–720
Xiaobo X, Jianping L, Peilin C (2006) Advances in the research and development of acrylic acid production from biomass. Chin J Chem Eng 14:419–427
Zhang J-F, Liu Z-Q, Zheng Y-G (2013) Improvement of nitrilase production from a newly isolated Alcaligenes faecalis mutant for biotransformation of iminodiacetonitrile to iminodiacetic acid. J Taiwan Inst Chem Eng 44:169–176
Acknowledgments
This work was supported by the National Key Basic Research Project 973 (2013CB733600) and the National Natural Science Foundation of China (No. 21476126).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, J., Yu, H., Chen, J. et al. Ammonium acrylate biomanufacturing by an engineered Rhodococcus ruber with nitrilase overexpression and double-knockout of nitrile hydratase and amidase. J Ind Microbiol Biotechnol 43, 1631–1639 (2016). https://doi.org/10.1007/s10295-016-1840-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1840-9