Abstract
The present work is aimed to examine the genetic variability and the distribution pattern of beneficial Trichoderma spp. isolated from rhizosphere samples and their mode of action in improving the plant health. A total of 131 suspected fungi were isolated from the rhizospheric soil and 91 isolates were confirmed as Trichoderma spp. T. asperellum and T. harzianum were found high in the frequency of occurrence. Genetic diversity analysis using RAPD and ISSR revealed the diverse distribution pattern of Trichoderma spp. indicating their capability to adapt to broad agroclimatic conditions. Analysis of genetic diversity using molecular markers revealed intra-species diversity of isolated Trichoderma spp. The frequency of pearl millet (PM) root colonization by Trichoderma spp. was found to be 100%. However, they showed varied results for indole acetic acid, siderophore, phosphate solubilization, β-1,3-glucanase, chitinase, cellulase, lipase, and protease activity. Downy mildew disease protection studies revealed a strong involvement of Trichoderma spp. in direct suppression of the pathogen (mean 37.41) in the rhizosphere followed by inducing systemic resistance. Our findings highlights the probable distribution and diversity profile of Trichoderma spp. as well as narrate the possible utilization of Trichoderma spp. as microbial fungicides in PM cultivation across different agroclimatic zones of India.
Similar content being viewed by others
Introduction
Pearl millet (Pennisetum glaucum (L.) R. Br.) (PM) is one of the most significant millet cultivated across semi-arid regions of the world. India is one of the largest producers of PM in the world, with approximately 7 million ha (Mha) area under cultivation. Rajasthan is the highest PM producing state within the country followed by Maharashtra, Gujarat, Uttar Pradesh, and Haryana, it accounts for more than 90% of PM acreage in the country. PM is the fourth most extensively cultivated crop after rice, wheat, and maize in India. It occupies an area of 6.93 Mha with an average production of 8.61 million tonnes with a productivity of 1243 kg/ha during 2018–20191. It is the staple diet for the poorest people, and its stover is used as livestock feeds in rural India. As PM cultivation is carried out on marginal land, its productivity is always lesser than expected. Additionally, continuous application of synthetic fertilizers and pesticides in this region leads to reduced soil health by decreasing the population of native microbes and releasing organic carbon2,3. On the other hand, downy mildew (DM) of PM caused by Sclerospora graminicola is a widespread devastating disease reported to cause substantial annual yield losses in India4,5,6. The annual grain yield loss due to DM is estimated to 20–40%, however, the losses are much higher under unfavorable conditions of high relative humidity, moderate temperature, and extensive use of the same cultivar7,8. DM disease has been managed conventionally by seed priming with fungicide Apron 35 SD and foliar spray with Ridomil MZ9.
Agriculture relatively relies on close associations between plants and the microbes that live in association with plant roots. These rhizosphere microorganisms are distinct from other microbial communities with their high biocontrol potential and they are more important to the plant for nutrient uptake and to boost plant health against pathogen10. To achieve this, it is important to ascertain the functional and structural diversity of desired microbes within particular agroclimatic conditions. Trichoderma has been widely employed and studied as a biocontrol agent against a wide range of phytopathogens including bacteria11,12, fungi13,14,15,16, oomycetes17,18, and nematode19,20 in a wide range of crops and climatic conditions. In India, approximately 250 commercial formulations of Trichoderma spp. are being used21,22, and identification and characterization of new species for their potential biocontrol properties are under progress23. Trichoderma can reduce the severity of plant diseases either by directly inhibiting the growth and establishment of phytopathogens or by inducing systemic resistance (ISR) in the host plant22,24,25. Mechanism suggested to be involved in the biocontrol activity of Trichoderma spp. are antibiosis, lysis, competition, and mycoparasitism26,27. Trichoderma are capable to activate both SAR and ISR mechanism in plant hosts28. Trichoderma spp. are identified for their ability to produce bioactive secondary metabolites, including polyketides, terpenoids, alkaloids and peptaibols29. SMs secreted by various Trichoderma spp. own the potential to inhibit the growth of important plant pathogenic microrganisms. T. harzianum control the growth of Clavibacter michiganensis subsp. michiganensis by producing Lysosime30. T. harzianum T23 inhibit the growth of Erwinia amylovora and C. michiganensis in vitro by producing viridiofungin A31. Different peptaibols such as Trichokonin VI, VII and AVIII were identified in Trichoderma spp., which are the cause of the suppression of plant pathogens growth32. Trichoderma spp. are known to produce enzymes like chitinase and glucanase which can degrade fungal and oomycetous pathogen cell walls and also reported to produce antibiotics33. Further, they are known to produce phytohormones such as gibberellic acid and indole acetic acid (IAA), and cycling soil nutrients, thereby playing a crucial role in enhancing soil organic carbon and improving the soil configuration and fertility. Together with all these properties of Trichoderma spp. are involved in improving plant growth and health34,35,36.
The prospect of influencing crop rhizospheric microbial inhabitants by inoculating useful microorganism(s) to enhance plant growth and health has revealed substantial assurance in greenhouse and laboratory studies; however, variable responses have been noticed in the field studies37. The main reason behind the reduced efficiency of biocontrol agents under field conditions is their ability to adapt to local biotic and abiotic environmental conditions. To understand this phenomenon, it is necessary to study the geographical distribution pattern of biocontrol agents in the rhizosphere. Hence, the present investigation was undertaken to study (1) the distribution and genetic diversity patterns of Trichoderma spp. in PM growing regions of India, (2) the characterization of Trichoderma for their beneficial traits, and (3) the potential of Trichoderma to improve the host plant growth and suppress DM disease in PM.
Results
Suspected 131 fungal were isolated from 193 soil samples collected from 58 districts distributed across eight agroclimatic zones (Supplementary Fig. S1). They were morphologically characterized by growing on PDA plates. The isolates recorded wide morphological characters. Colonies of Trichoderma spp. appear initially w.hitish wooly and become compact as they grow. During conidia formation, they turn blue-green or yellow-green color. Some isolates grow in the form of concentric rings. The reverse is pale greenish, tan, and yellowish in different Trichoderma spp. (Fig. 1). Further the phialides, conidial arrangement, and conidial morphology varied among the isolates. The identity of the isolates was further confirmed by analyzing the ITS sequence (Supplementary Table S1). Collectively, these isolates were identified as T. asperellum (35.1%), T. harzianum (27.5%), T. virens (16.5%), T. longibrachiatum (5.5%), T. atroviride (2.1%), T. brevicompactum (1.1%), T. viride (4.4%), T. hamatum (2.2%), and five isolates (5.5%) were only identified up to genus level (Fig. 1). Among 131 Trichoderma isolates, 91 species (after removing duplicates and other fungal isolates), which are decisively identified, based on morphological and molecular profiles were selected for further studies. ITS sequence of Trichoderma isolates was submitted to the National Center for Biotechnology Information (NCBI, Bethesda, Maryland, USA) database, and an accession number was obtained. Distribution analysis revealed that Trichoderma spp. are widely distributed across different agroclimatic regions of India. Among the identified isolates, T. asperellum and T. harzianum were found in all agroclimatic regions. Whereas, T. brevicompactum was recorded only in soil samples collected from the state of Uttar Pradesh (Supplementary Table S1).
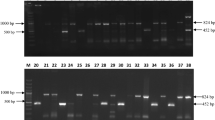
RAPD was carried out by using 20 sets of random primers. All the primers formed reproducible and polymorphic bands and were further selected for screening. In the size range of 0.1–5.0 kb, 97 bands were scored, with an average of 4.85 bands per primer. Among these bands, 51 were polymorphic in nature with 52.58% polymorphisms (Table 1). RAPD-7 seemed to be most proficient with 87.5% polymorphism compared to others. The PIC values were recorded in the range from 0.451 (RAPD-3) to 0.894 (RAPD-7) with average heterozygosity (mean of PIC) of 0.718. A representative illustration of RAPD profile obtained by RAPD-7 primer in the diversity studies of 91 Trichoderma spp. is shown in the supplementary Fig. S2.
In ISSR assays, 12 primers showed polymorphic and reproducible bands. From the 91 isolates of Trichoderma spp., 54 bands were scored in the size range of 0.1–5 kb, with an average of 4.5 bands per primer. Of these 54 bands, 23 were polymorphic with 42.59% polymorphism (Table 2, Supplementary Fig. S3). ISSR-5 was found to be most efficient with 75.0% polymorphism compared to other primers. The PIC values were observed in the range from 0.453 (ISSR-6) to 0.874 (ISSR-4) with mean heterozygosity (average of PIC) of 0.704. However, in the mantel test (Fig. 2), a weak correlation (R2 = 0.095) was observed among genetic distance analyzed by RAPD and ISSR data of the Trichoderma isolates. The Unweighted Pair-Group Methods with Arithmetic Average (UPGMA) cluster analysis among 91 Trichoderma isolates produced by RAPD and ISSR primers is represented in Fig. 3. Principle coordinated analysis (PCA) based on RAPD and ISSR pattern results revealed that these isolates were not a region or host-specific (Fig. 4), which indicated the distribution of Trichoderma isolates in wide agroclimatic zones and also its association with multiple host systems.
All the 91 Trichoderma isolates were screened for their beneficial traits. In the root colonization study, 100% positive results were noticed in all Trichoderma seed treatment under greenhouse conditions (Supplementary Fig. S4). In phosphate solubilization assay, 58.2% of Trichoderma isolates showed a strong ability to solubilize phosphate complex present in the culture media. In contrast, only 46.1% of isolates were found producing IAA. Among the 91 isolates tested for the production of lytic enzymes, 57.2% isolates were found positive for siderophore production, 82.4, 62.6, 64.8, and 57.1% isolates were found positive for chitinase, glucanase, cellulase, and protease activity, respectively. But, in all the above studies, a certain number of Trichoderma isolates recorded variable results (Fig. 5).
All the 91 Trichoderma spp. were screened for their ability to improve seed quality parameters of PM. PM seedlings were analyzed for fold increase or decrease in vigor index over control upon seed treatment with different Trichoderma spp. Among the 91 Trichoderma isolates, 53 isolates showed significantly (p ≤ 0.05) highest seedling vigor index. None of the treatments inhibited the normal growth of seedlings. It was recorded that the seedling vigor of primed seeds was significantly (p ≤ 0.05) higher on an average of four-fold increase over control. Seed treatment with Metalaxyl provided 88% germination of seeds and 1498 seedling vigor, which were not significantly (p ≤ 0.05) different from the control (Fig. 6).
It was recorded that the disease protection ability of Trichoderma spp. was significantly (p ≤ 0.05) higher (mean 37.41) when the pathogen was inoculated only as soil inoculation. However, the combined application of pathogen (soil and whorl inoculation) significantly (p ≤ 0.05) decreased the disease protecting ability of Trichoderma (mean 28.78). Control treatment with sterile distilled water (SDW) showed maximum disease incidence of 95.6% (soil treatment) and 93.8% (soil treatment + whorl inoculation) (Fig. 7).
Effect of seed priming with T. asperellum (DL-81) formulation was evaluated in DM sick plot under epiphytotic field conditions. Maximum protection with least incidence was observed in T. asperellum (DL-81) treated seedlings with maximum protection with least disease incidence, but in the case of the control treated plot, the highest disease incidence i.e., 96% was noticed and had not shown any protection against DM pathogen in the host plant (Fig. 8). The seed quality parameter of T. asperellum treatment on growth parameters of PM plants under field conditions was summarized in Table 3. Chemical metalaxyl treatment exhibited maximum of 86% disease protection.
Discussion
In this study, we intended to understand the distribution patterns of Trichoderma across different agroclimatic zones, functional diversity, and about beneficial interaction with plants and possible applications to improve the growth and health of PM. Distribution of Trichoderma is governed by several factors which include cropping pattern38, root exudates39,40,41, water availability, low osmotic levels42,43, soil pH44,45, temperature46, metal ions47, pesticides48, other biotic and abiotic stress conditions. In spite of this, Trichoderma is known to adapt to variable conditions and survive by imposing its beneficial effects on the local environment. These observations are encouraging in the sense that these fungi can be used to develop bioformulations, which can be applied in multiple agroclimatic conditions and also to different plant host systems.
Though more than 200 species of Trichoderma were reported across the world, including India. In the present study, we have observed only nine species predominantly associated with PM, wheat, and maize. Previously, Blaszczyk et al.49 reported fourteen Trichoderma spp. among which, T. harzianum was found to be the most abundant in Poland. In South Africa, Du-Plessis et al.50 recorded fourteen different species of Trichoderma among which ten were for the first time reported in this region. In Manipur (India), 22 different species of Trichoderma were identified from nine geographically diverse zones. They were diverse in their phylogeny, and T. harzianum was most predominant in occurrence51. In the present investigation, Trichoderma isolates exhibited significant intra-species and inter-species genetic variations as observed through RAPD and ISSR analysis.
With evidence from the similar average values of PIC (0.718 for RAPD and 0.704 for ISSR), RAPD and ISSR based assays were found equally useful for the analysis of genetic diversity of isolated Trichoderma spp. Also, Mantel test reveals the weak correlation (R2 = 0.095) among genetic distance analyzed by RAPD and ISSR data. This indicates that these two sets of markers target different regions of the Trichoderma genome. Similar results of low levels of correlation between RAPD and ISSR markers were observed in microbes and higher plants52.
In the present study, the UPGMA cluster analysis based on RAPD and ISSR data, studied 91 Trichoderma isolates were not clustered according to their place of collection or host plant (Fig. 4). This may be due to the high genetic diversity and wide spread of Trichoderma spp. across the India. Moreover, in the UPGMA cluster, AP-42 isolate (T. harzianum, collected from Guntur, Andhra Pradesh, India, from the host plant PM), followed by BG-21 (T. longibrachiatum, collected from Bagalkote, Karnataka, India, from the host plant maize) were grouped distantly separated from the other accession, suggesting the lowest genetic identity of these accessions with others.
The PCA plot constructed from RAPD and ISSR binary matrix showed the substantial distribution of Trichoderma spp. collected from the different agroclimatic regions (Fig. 5). Moreover, the wide distribution of Trichoderma spp. in the PCA plot revealed the existence of significant genetic diversity. The presence of considerable bio-diversity of Trichoderma spp. from soils of the particular region offers opportunities for further sustainable agriculture management practices, especially in the progress of identifying the bio-control agent to manage plant diseases53.
New-fangled haplotypes have been documented in T. harzianum based on the study of ITS sequences phylogenetic analysis and were substantiated by data on RAPD profile54. Samuels et al.55 differentiated T. aggressivum f. aggresivum and T. aggressivum f. europaeum from morphologically parallel species based on temperature growth associations, examined the aptitude to cause green mold disease of mushrooms, and further analyzed the ITS-1 region, elongation factor gene sequence. Hermosa et al.56 studied the genetic diversity of 69 biocontrol isolates of Trichoderma spp. from different geographical regions and culture collections based on the examination of sequence data attained from the ITS1 province and a segment of the tef1 gene. Growth promoting and beneficial traits of Trichoderma isolates are highly correlated and found varying from isolate to isolate. It was observed that chitinase uses an essential mechanism of pathogen suppression and also IAA production and phosphate solubilization is important for plant growth improvement. These benefical traits are extensively studied and correlated with plant growth, health, biomass, and yield of the plant57,58. Phosphate solubilization, phytohormone production, lytic enzymes, and antibiotic production are the main characteristics of Trichoderma in promoting plant growth and biocontrol activity59,60.
In general, the mechanism involved in plant disease suppression by Trichoderma is identified as, directly through antagonism or indirectly by elicitation of plant-defense responses61,62. Disease protection by direct suppression is through mycoparasitism63,64 fabrication of lytic enzymes such as chitinase65, β-1, 3 glucanase66,67, protease68, and antimicrobial chemicals (Trichoharzianol, Isoharziandione, 6-pentyl-α-pyrone)69,70,71 and competing for space and food. Disease protection by ISR has been reported in several plant disease managements by Trichoderma spp. including PM-DM72,73.
Cell wall glucan of T. hamatum UOM 13 was found to suppress the DM diseases in PM by inducing systemic resistance74. Further Siddaiah et al.73 reported that the UOM 13 induced resistance was through enhanced structural defense (lignification and callose deposition), upregulation of defense-related enzymes (Glucanase, Peroxidase, Phenylalanine ammonia-lyase, Polyphenol oxidase), PR protein, and Hydroxyproline-Rich Glycoproteins in challenged PM seedlings. Involvement of β-1,3-glucanase in suppressing the DM disease was demonstrated by O'Kennedy et al.75 were the genes of β-1,3-glucanase (gluc78) from T. atroviride, was introduced into the genome of pearl millet (842B), by particle bombardment and expressed. Further, a clear correlation was established between increased β-1,3-glucanase activity and decreased DM disease in transgenic PM seedlings. In our earlier studies18,76 the capabilities of Trichoderma crude protein and lipid fractions of T. brevicompactum (UP-91) to induce systemic resistance against DM infection was observed. Further, the active lipid molecule involved in ISR is suspected as (E)-N-(1,3-dihydroxyoctadec-4-en-2-yl) acetamide. The colonization of the rhizosphere by Trichoderma spp. produces direct positive effects on plants, promoting their growth, and activating their defense mechanisms. T. asperellum and T. atroviride modulated salicylic acid, jasmonic acid, and antifungal defensin pathways in tomato plants during the early stage of the root pathogen infection77. T. harzianum ETS323 secrets an flavoenzyme, L-amino acid oxidase has antimicrobial characteristics by activating H2O2 signaling defense mechanisms to confer resistance against S. sclerotiorum and B. cinerea in tobacco78.
We have studied S. graminicola, an oomycetous pathogen in PM host plant, and its influence and modulation on biochemical parameters upon seed priming with Trichoderma spp. As the major cell wall constituents of oomycetes are β-1, 3-glucan79, it is believed that glucanase will play a crucial role in suppressing DM pathogen over chitinase. In the present study, most of the Trichoderma isolates recorded significant (p ≤ 0.05) reduction in DM incidence, when the pathogen was inoculated in the soil. Though, the detailed disease protection mechanisms were not analyzed in the current study. It was presumed that it follows various mechanisms mentioned above. However, their disease protection ability was found to decrease after providing an additional whorl inoculation of the pathogen. This observation revealed that the principal mechanism with which Trichoderma reduces the disease is by suppressing pathogen by directing inhibition. However, their ability to ISR varies among isolates. The present study showed the current status of Trichoderma distribution in different PM growing agroclimatic regions of India. Moreover, the wide occurrence of nine Trichoderma spp. beyond the barriers of environmental conditions and the host system enhanced the probability of employing these bioagents for the improvement of PM growth and health.
Methods
Isolation, identification, and characterization of Trichoderma spp
To isolate Trichoderma spp., field surveys were conducted under different agroclimatic zones of India, which includes 10 different states (Karnataka, Tamil Nadu, Andhra Pradesh, Madhya Pradesh, Maharashtra, Rajasthan, Gujarat, Haryana, Uttar Pradesh, and Delhi) belonging to eight different agroclimatic zones (Supplementary Fig. S1), in between September 2012 to January 2014. In each field, rhizosphere soils of five randomly selected healthy looking plants were collected and pooled to obtain a composite sample. Soil samples were collected into sterile cotton bags, transported to the laboratory, and immediately processed.
Rhizospheric soil (10 g) was mixed with 250 ml of phosphate-buffered saline containing streptomycin (100 mg/l) and penicillin (100 mg/l) to suppress the native bacterial growth and incubated at 28 ± 2 °C for 2 h on a rotary shaker at a speed of 150 rpm. Ten-fold serial dilutions were prepared and from each dilution, 100 μl were inoculated by spread plate method on Martin medium80 amended with streptomycin (100 mg/l) and penicillin (100 mg/l). The inoculated plates were incubated for 7 days at 28 ± 2 °C and visually observed for the fungal growth at regular time intervals. At the end of the incubation period, the suspected fungus was isolated onto potato dextrose agar (PDA) slants.
The fungal isolates were identified based on their morphological nature by growing them on PDA at 28 ± 2 °C for 5–7 days. The fungal colonies were visually observed for their color (obverse and reverse), texture, margin, and sporulation. Individual fungal isolates were identified at the species level using morphological keys and species descriptions (conidiophores, shape, size, arrangement, and phialides arrangements were analyzed microscopically) as per the protocol suggested by Rifai81 and Leahy et al.82. Fungal strains were sub-cultured and grown on PDA for routine experiments and the long-term storage, fungal slants were prepared in cryovials overlaid with 20% glycerol and stored at − 80 °C.
Genomic DNA was extracted following the method of Saghai-Maroof et al.83. PCR amplification of ITS region was performed using forward ITS1 (5′ TCC-GTA-GGT-GAA-CCT-GCG-G 3′) and reverse ITS4 (5′ TCC-TCC-GCT-TAT-TGA-TAT-GC 3′) primers following the conditions given by White et al.84. Further, the terminal regions of DNA sequence with deprived alignment were manually detached via BioEdit v 7.0.5.3 and subjected to BLAST searches to consign putative identity, the depiction of operational taxonomic entities based on phylogenetic implication and sequence similarity measures. The sequences were deposited in the NCBI database and an accession number was obtained (Supplementary Table S1).
Molecular Phylogenetic analysis and evolutionary history were grouped based on the Tamura-Nei model by using the Maximum Likelihood method85. The bootstrap consensus tree was inferred from 1000 replicates taken to symbolize the evolutionary history of the taxa examined using MEGA686,87.
Genetic diversity analysis
A set of 20 RAPD primers and 12 ISSR primers were used for the polymorphism study (Table 2). RAPD analysis was performed in 0.2 ml PCR vials including reaction buffer, 1 µl of dNTP mix (2.5 M each), 2.5 µl of 10 × PCR buffer, 1U Taq DNA polymerase, 30 ng DNA template, 10 pmol of 1 µl primer, and DNase free water to a final volume of 25 µl. The amplification of the PCR reaction was carried out at 94 °C for 3 min (initial denaturation), followed by 45 cycles of 94 °C for 1 min (denaturation); 40 °C for 1 min (annealing temperature of primer); 72 °C for 2 min (extension of primer) and 72 °C for 10 min (final extension).
For the ISSR analysis, annealing temperature differs for the specific primer based on Tm (Table 3), and other conditions were the same as explained for RAPD. PCR amplified products were separated and analyzed on the agarose gel. The sizes of the amplicon were calculated with a DNA ladder of 100–5000 bp and imaged using a gel documentation system (BioRad, Canada). For each DNA sample, the process of RAPD and ISSR analysis was performed twice and primers with reproducible nature were subjected to diversity analysis.
The amplified products of different sizes were scored as 1 (presence) and 0 (absence) for 91 Trichoderma spp. to produce a binary matrix. Analysis of data was carried out using the software NTSYS-pc, version 2.11w (Numerical Taxonomy System Biostatistics) to analyze the Jaccard’s similarity coefficients88. The SIMQUAL program was employed to estimate the Jaccard’s coefficients. The dendrogram was produced by grouping Jaccard coefficients using the Sequential agglomerative hierarchical non-overlapping (SAHN) clustering program, choosing the algorithm of Unweighted Pair-Group Methods with Arithmetic Average (UPGMA) in NTSYS-pc89.
Polymorphic information content (PIC) was studied using the method: PIC = 1 − ∑pi2, where ‘pi’ is the frequency of the ith allele90. Percent polymorphism was calculated according to the principle: % Polymorphism = p/(m + p), where p is a sum of the number of polymorphic bands and m is the total number of monomorphic bands of the primer alliance used. Multiplex ratio (MR) for all markers was determined via the formula: MR = (m + p)/n, where p is the total number of polymorphic bands, m is the total number of monomorphic bands and n is the total number of primer combinations91. Average heterozygosity (Hav) was known by taking the average of PIC values acquired for all the markers. Marker index (MI) was studied by multiplying the Hav with the MR. Genetic diversity of the 91 Trichoderma spp. and its interpretation was performed by the method explained by Saini et al.52.
Characterization of Trichoderma spp. for their beneficial traits
Root colonization bioassay
Seeds of PM (cv. 7042S) which was reported as highly susceptible to DM pathogen were obtained from the International Crop Research Institute for Semi-Arid Tropics, Patencheru, India. Sodium hypochlorite (0.2%) was used for the surface sterilization of seeds, later thoroughly washed with SDW, and used throughout the experimental studies. Pure cultures Trichoderma spp. were obtained by growing on PDA. 10 ml SDW was added to PDA plates and conidia were dislodged from the culture surface and the concentration was adjusted to 1 × 108 conidia/ml using a hemocytometer.
PM seeds were treated with a conidial suspension of Trichoderma spp. amended with 0.2% Carboxymethylcellulose (CMC) as adhesive for 3 h at 28 ± 2 °C on a rotary shaker (150 rpm). Seeds primed only with SDW amended with CMC were referred to as control. Treated seeds were sown thickly in pots (22 cm diameter) filled with the sterilized potting mixture [soil: farmyard manure: sand (2:1:1, v/v/v)]. The earthen pots with different treatments were randomly arranged in block designs and maintained under greenhouse conditions at 25–30 °C with 95% relative humidity (day-night light cycle consisting of an average temperature and light of 18 ± 2 °C in the night and 28 ± 2 °C in the day with natural light). After 7 days, the seedlings were thinned to space out by leaving six to eight seedlings per pot. After 21 days of incubation, plants were carefully removed without damaging the roots and the rhizosphere soil was collected. The soil was processed as explained above and the colony-forming unit (cfu) was determined following the serial dilution technique on Martin’s media. Visual and microscopic observations were carried out as explained earlier. For further confirmation, root bits (3 mm) were placed on Martin media, and the fungal growth was analyzed. All bioassays were performed in triplicates of six pots each and repeated twice.
Indole acetic acid production
Trichoderma spp. were grown in potato dextrose broth (PDB) amended with L-Tryptophan (0.1 g/L) for 4–5 days at 28 ± 2 °C on a rotary shaker at 150 rpm. At the end of the incubation period, mycelia and debris were separated by filtration and centrifugation, respectively. To 1 ml of the culture filtrate, 4 ml of Salkowski reagent (2% 0.5 M ferric chloride in 35% perchloric acid) was added, and development of pink to red color was visually observed after 10 min of the incubation period in dark condition92. An uninoculated PDB was maintained as a control. For each isolate, the experiment was conducted in triplicates and repeated thrice.
Phosphate solubilization
Calcium phosphate solubilization ability of different Trichoderma spp. was determined by growing them on National Botanical Research Institute’s Phosphate growth medium (NBRIP)93. The petriplates containing NBRIP was point-inoculated and incubated at 28 ± 2 °C for 4–5 days. Change in color of the media surrounding or beneath the fungal media from purple to yellow indicates the ability of an isolate to solubilize calcium phosphate.
Siderophore production
Trichoderma isolates were point inoculated onto Chrome azurol S (CAS) media and incubated for 5–7 days at 28 ± 2°C94. Development of pink or yellow or orange coloration around or below the fungal colony indicates the positive nature of fungal isolate to produce siderophore.
Lytic enzyme assay
The Trichoderma isolates were grown on 1/10th strength PDB for 48 h on a rotary shaker at 150 rpm at 28 ± 2 °C. For induction of enzyme production, the specific substrate (CMC for cellulase; colloidal chitin for chitinase; laminarin for β, 1–3, glucanase and casein for protease) was added at 0.5% (w/v) into the culture medium. Further, the incubation was continued for 24 h, and culture filtrate was separated by filtration and centrifugation (7800g; 4 °C; 10 min) and used as an enzyme source.
Chitinase (EC 3.2.1.14) activity was quantified spectrophotometrically. Briefly, 100 µl of the culture filtrate was blended with 200 µl of colloidal chitin (0.5% in 50 mM acetate buffer pH 5.2). The reaction was incubated for 60 min at 40 °C with intermediate shaking. To stop the reaction, the vials were placed in a boiling water bath for 5 min. After cooling, 3 ml of freshly prepared dimethylaminobenzaldehyde solution was added and incubated at 40 °C for 60 min. A standard graph was prepared using different concentrations of N-Acetylglucosamine (GLcNAc) and the enzyme activity was measured by quantifying the released GLcNAc95.
β-1,3 glucanase (EC 3.2.1.39) activity was evaluated by adding100 µl culture supernatant to 200 µl of laminarin (5%, w/v) prepared in 50 mM acetate buffer (pH 5) and further incubated at 40 °C for 10 min. Reducing sugar was measured by the 3,5-dinitrosalicylic acid (DNS) method96.
Cellulase activity (EC 3.2.1.21) was examined by adding 100 µl of the culture supernatant to 400 µl of CMC (1%) prepared in 100 mM sodium citrate buffer (pH 5.2) and the assay mixture was further incubated at 55ºC for 15 min. The reducing sugars released were determined by the DNS technique96.
To analyze Protease (EC 3.4.21.4) activity, 500 µl of the culture supernatant was added to 500 µl of casein (0.36%) and 2 ml of acetate buffer (100 mM, pH 3.6). Towards the end of the reaction period (1 h, 50ºC), 3 ml of trichloroacetic acid (5%) was added to stop the reaction. Blank was considered a zero period of incubation. Free amino acids released were measured by the Ninhydrin method97.
The concentration of proteins in the samples were verified using the Bradford method98, with bovine serum albumin (BSA; Sigma Aldrich, St. Louis, USA) as a standard. All enzyme assays were replicated three times for each sample.
The specific activity was articulated as Unit mg−1 protein and Unit mg−1 aminoacid (for protease). The activity is described as the quantity of enzyme required to generate one mM of equivalent reducing sugar in one min of one ml culture supernatants. Non-enzymatic controls were also carried out using boiled enzymes and were deducted from the enzymatic values.
Effect of Trichoderma seed treatment on seed quality variables of pearl millet
Seed treatment was conducted aseptically as explained earlier in the laminar airflow and the seeds were dried overnight and used. Seed germination was tested following the paper towel method99. Mean root length (MRL) and Mean shoot length (MSL) were measured and seedling vigor was calculated using the following formula100.
For each treatment, four samples of 100 seeds were used, and the experiment was replicated thrice.
Disease protection studies
Sclerospora graminicola sick plot is being maintained in the University of Mysore at Department of Biotechnology, Mysuru, for the last 30 years under the Indian Council of Agricultural Research (ICAR)-All India Coordinated Pearl Millet Improvement Project (AICPMIP, Jodhpur Rajasthan, India). The pathogen was isolated from the highly susceptible PM leaves grown in the sick plot. Infected leaves were cleaned with running tap water to eliminate debris and existing sporangia, blot-dried, and incubated in a moist chamber at 80% humidity at 20 °C under dark conditions. Fresh sporangia formed on the infected leaves were harvested in SDW, and spore concentration was adjusted to 4 × 104 per ml using a hemocytometer and utilized as inoculum101. Oospores were obtained from the infected leaves of pearl millet which was containing approximately 5 × 105 oospores per gram dry weight.
Greenhouse conditions and the methods of the pot experiment were performed as explained earlier. Disease protection studies were conducted in two sets of experiments, (1) control and treated seeds were raised in sterile soil and pathogen (zoospores) was challenge inoculated as whorl inoculation102 and (2) control and treated seeds were raised in soil containing pathogen (~ 5 × 105 oospores) and an additional whorl inoculation was performed as explained above. In both cases, the challenge-inoculated plants were maintained under greenhouse conditions (80% RH). Disease frequency was monitored by examining the number of plants that confirmed any one of the characteristic DM symptoms (chlorosis, sporulation on the abaxial leaf surface, stunted growth, etc.) up to 30 days after sowing. For each Trichoderma isolate, eight pots of two replicates were maintained, and the experiment was repeated twice.
Trichoderma treatment offering the best protection under greenhouse conditions against DM disease was selected for the field trials. Field studies were conducted in the DM sick plot of DOS in Biotechnology, University of Mysore (N24° 18′, E79° 26′, 903 m altitude, red loam soil). This experimental station has been maintained over three decades under the ICAR program, with a severe infestation of the DM oospores inoculum and additional inoculum in the form of asexual spores is provided from spreader rows raised 21 days before sowing of experimental plot103. The treated seeds were sown in randomized block design 10 × 6 m plots with four replications per treatment. Each row was 6 m long and 75 cm apart with 15 cm spacing between plants within rows. Susceptible controls were similar as described earlier. The plants were raised following recommended agronomical practices. Disease incidence was recorded at 30 and 60 days growth of seedlings, as the plants started showing typical DM disease symptoms.
Plant growth promotion and yield analysis under field conditions
Seed treatment with T. asperellum (DL-81) and seedlings were raised in the field as explained above. Except, these set of experiments were conducted in DM disease-free plots and the seedlings were not subjected to challenge inoculation with pathogen. Towards the end of the growth period (60 days), plant height (cm), Number of productive ear head, Length of ear head (cm), Girth of ear head (cm), and 1000-seed weight (g) was measured and tabulated.
Statistical analysis
All the data from laboratory and greenhouse experiments were subjected to one-way analysis of variance (ANOVA) using SPSS, version 17. The significant difference between the averages of treatments were compared using Highest Significant Difference (HSD) as attained by Tukey test at p ≤ 0.05 levels.
Ethical approval
All authors concur with the submission and have seen a draft copy of the manuscript and agree with its publication. The manuscript does not contain experiments using animals and human studies.
References
Satyavathi, C. T. Project Coordinator review, 55th Annual Group Meeting, 13–15th March 2020. ICAR – All India Coordinated Research Project on Pearl millet, Jodhpur, Rajasthan – 342304, India (2020). Website: http://www.aicpmip.res.in; http://aicrp.icar.gov.in/pearl.
Kaur, K., Kapoor, K. K. & Gupta, A. P. Impact of organic manure with and without mineral fertilizers on soil chemical and biological properties under tropical conditions. J. Plant Nutr. Soil Sci. 168, 117–122 (2005).
Bitew, Y. & Alemayehu, M. Impact of crop production inputs on soil health: A review. Asian J. Plant Sci. 16, 109–131 (2017).
Singh, S. D., King, S. B. & Werder, J. Downy mildew disease of pearl millet. Information Bulletin no. 37. International Crops Research Institute for the Semi-Arid Tropics. Patancheru, Andhra Pradesh, India. pp 36 (1993)
Thakur, R. P., Shetty, H. S. & Khairwal, I. S. Pearl millet downy mildew research in India: progress and perspectives. Int. Sorghum Millets Newslett. 47, 125–130 (2006).
Thakur, R. P. Pearl millet Disease management in arid land crops. In Scientific Publishers (eds Lodha, S. et al.) 21–41 (Jodhpur, 2008).
Singh, S. D. & Singh, G. Resistance to pearl millet hybrid NBH 3. Ind. Phytopathol. 40, 178–180 (1987).
Thakur, R. P. et al. Evidence for a new virulent pathotype of Sclerospora graminicola on pearl millet. Indian J. Mycol. Plant Pathol. 29, 61–69 (1999).
Prakash, H. S., Nayaka, C. S. & Kini, K. R. Downy mildew disease of pearl millet and its control. In Future challenges in crop protection against fungal pathogens (eds Goyal, A. & Manoharachary, C.) 109–129 (Fungal Biology, Springer, 2014).
Schmidt, J. E., Vannette, R. L., Igwe, A., Blundell, R., Casteel, C. L. & Gaudin, A. C. M. Effects of agricultural management on rhizosphere microbial structure and function in processing tomato plants. Appl. Environ. Microbiol. 1, 85(16), e01064–19 (2019).
Leelavathi, M. S., Vani, L. & Reena, P. Antimicrobial activity of Trichoderma harzianum against bacteria and fungi. Int. J. Curr. Microbiol. Appl. Sci. 3, 96–103 (2014).
Anwar, J. & Iqbal, Z. Effect of growth conditions on antibacterial activity of Trichoderma harzianum against selected pathogenic bacteria. Sar. J. Agri. 33, 501–510 (2017).
Papavizas, G. G. Trichoderma and Gliocladium: Biology, ecology and potential for biocontrol. Ann. Rev. Phytopathol. 23, 23–54 (1985).
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I. & Lorito, M. Trichoderma species–opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56 (2004).
Babu, K. N. & Pallavi, P. N. Isolation, identification and mass multiplication of Trichoderma an important bio-control agent. Int. J. Phar. Life Sci. 4, 2320–2323 (2013).
Motlagh, M. R. S. & Samimi, Z. Evaluation of Trichoderma spp., as biological agents in some of plant pathogens. Ann. Biol. Res. 4, 173–179 (2013).
Nandini, B., Hariprasad, P., Niranjana, S. R., Shetty, H. S., & Geetha, N. P. Elicitaion of resistance in pearl millet by oligosaccharides of Trichoderma spp. against downy mildew disease. J. Plant Inter. 8, 45–55 (2013).
Nandini, B., Hariprasad, P., Shankara, H. N., Prakash, H.S. & Geetha, N. Total crude protein extract of Trichoderma spp. induces systemic resistance in pearl millet against the downy mildew pathogen. 3 Biotech 7, 183(2017).
Meyer, S. L. F. et al. Application of Burkholderia cepacia and Trichoderma virens, alone and in combinations, against Meloidogyne incognita on bell pepper. Nematropica 31, 75–86 (2001).
LaMondia, J. A. & Cowles, R. S. Effects of entomopathogenic nematodes and Trichoderma harzianum on the strawberry black root rot pathogens Pratylenchus penetrans and Rhizoctonia fragariae. J. Nemat. 34, 351–357 (2002).
Singh, H. B., Singh, B. N., Singh, S. P. & Sarma, B. K. Exploring different avenues of Trichoderma as a potent bio-fungicidal and plant growth promoting candidate: An overview. Rev. Plant Pathol. 5, 315–426 (2012).
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M. & Kenerley, C. M. Trichoderma research in the genome era. Ann. Rev. Phytopath. 51, 105–129 (2013).
Kredics, L., et al. Biodiversity of the genus Hypocrea/Trichoderma in different habitats. In: Gupta, V. G. et al. (ed) Biotechnology and Biology of Trichoderma. 3–24 (2014).
Viterbo, A., & Horwitz, B. A. (2010) Mycoparasitism In: Borkovich KA, Ebbole DJ (ed) Cellular and molecular biology of filamentous Fungi, Washington, Am. Soc. Microbiol. 42, 676–693.
Abdelrahman, M., Abdel-Motaal, F., El-Sayed, M., Jogaiah, S., Shigyo, M., Ito, S., & Tran. L. S. Dissection of Trichoderma longibrachiatum-induced defense in onion (Allium cepa L.) against Fusarium oxysporum f. sp. cepa by target metabolite profiling. Plant Sci. 246, 128–138 (2016).
Henis, Y. Biological control. In: Current Perspectives in Microbial Ecology (ed.) Klug, M. J. & Reddy, C. A. Washington DC: American Society for Microbiology pp. 353–361 (1984).
Lynch, J. M. Fungi as antagonists. In: "New directions in biological control: alternative for suppressing agricultural pests and diseases". Liss, New York. 243–253 (1990).
Tucci, M., Ruocco, M., De Masi, L., De Palma, M. & Lorito, M. The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12, 341–354 (2011).
Chavez, J. R. et al. Prealamethicin F50 and related peptaibols from Trichoderma arundinaceum: Validation of their authenticity via in situ chemical analysis. RSC Adv. 7, 45733–45741 (2017).
Utkhede, R. & Koch, C. Biological treatments to control bacterial canker of greenhouse tomatoes. Biocontrol 49, 305–313 (2004).
El-Hasan, A., Walker, F., Schöne, J. & Buchenauer, H. Detection of viridiofungin A and another antifungal metabolites excreted by Trichoderma harzianum active against different plant pathogens. Eur. J. Plant Pathol. 124, 457–470 (2009).
Xiao-yan, S. et al. Broad-spectrum antimicrobial activity and high stability of Trichokonins from Trichoderma koningii SMF2 against plant pathogens. FEMS. Microbiol. Lett. 260, 119–125 (2006).
Vinale, F. et al. A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72, 80–86 (2008).
Shoresh, M., Harman, G. E. & Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Ann. Rev. Phytopathol. 48, 21–43 (2010).
Hermosa, R., Viterbo, A., Chet, I. & Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25 (2012).
Sayed, E. M. M., El-Hashash, M. M., Mohamed, H. R. & Abdel-Lateef, E. S. Phytochemical investigation and in vitro antioxidant activity of different leaf extracts of Salix mucronata Thunb. J. Appl. Pharm. Sci. 5, 080–085 (2015).
Bowen, G. D. & Rovira, A. D. The rhizosphere and its management to improve plant growth. Adv. Agron. 66, 1–102 (1999).
Celar, F. Influence of root exudates of different plant seedlings on mycelial growth of antagonistic fungi Trichoderma species and Gliocladium roseum. Zb. Biotech. Fak. Univ. Ljublj. Kmet. 79, 343–348 (2002).
Fernandez, E., Trillas, M. I. & Segarra, G. Increased rhizosphere populations of Trichoderma asperellum strain T34 caused by secretion pattern of root exudates in tomato plants inoculated with Botrytis cinerea. Plant Pathol. 66, 1110–1116 (2017).
Vishwakarma, K. et al. Exploring the role of plant-microbe interactions in improving soil structure and function through root exudation: a key to sustainable agriculture. In Plant-microbe interactions in agro-ecological perspectives (eds Singh, D. et al.) 467–487 (Springer, 2017).
Lombardi, N. et al. Root exudates of stressed plants stimulate and attract trichoderma soil fungi. Mol. Plant Micr. Inter. 31, 982–994 (2018).
Eastburn, D. M. & Butler, E. E. Effects of SOIL MOISTURE AND TEMPERATURE ON THE SAPROPHYTIC ABILITY OF Trichoderma harzianum. Mycologia 83, 257–263 (1991).
Badham, E. R. Growth and Competition between Lentinus edodes and Trichoderma harzianum on Sawdust Substrates. Mycologia 83, 455–463 (1991).
Delgado-Jarana, J., Pintor-Toro, J. A. & Benı́tez, T. ,. Overproduction of β-1,6-glucanase in Trichoderma harzianum is controlled by extracellular acidic proteases and pH. Biochem. Biophys. Acta. 1481, 289–296 (2000).
Muniappan, V. & Muthukumar, T. Influence of crop species and edaphic factors on the distribution and abundance of Trichoderma in Alfisol soils of southern India. Acta Bot. Croat. 73, 37–50 (2014).
Antal, Z., Manczinger, L., Szakács, G., Tengerdy, R. P. & Ferenczy, L. Colony growth, in vitro antagonism and secretion of extracellular enzymes in cold-tolerant strains of Trichoderma species. Mycol. Res. 104, 545–549 (2000).
Morley, G. F. & Gadd, G. M. Sorption of toxic metals by fungi and clay minerals. Mycol. Res. 99, 1429–1438 (1995).
Sesan, T. E. & Oprea, M. The in vitro action of fungicides and insecto-fungicides on the antagonistic fungi used as biocontrol agents. Bull. Pol. Acad. Sci. Tech. Sci. 47, 183–195 (1999).
Błaszczyk, L. et al. Species diversity of Trichoderma in Poland. J. App. Genet. 52, 233–243 (2011).
Du Plessis, I. L., Druzhinina, I. S., Atanasova, L., Yarden, O. & Jacobs, K. The diversity of Trichoderma species from soil in South Africa, with five new additions. Mycologia 110, 559–583 (2018).
Kamala, T., Devi, S. I., Thingnam, G. & Somkuwar, B. G. Genetic diversity and species pattern of Trichoderma and Hypocrea in Manipur using in silico analysis. Bioinformation 9, 106–111 (2013).
Saini, R. K., Saad, K. R., Ravishankar, G. A., Giridhar, P. & Shetty, N. P. Genetic diversity of commercially grown Moringa oleifera Lam. cultivars from India by RAPD, ISSR and cytochrome P450-based markers. Plant Sys. Evol. 299, 1205–1213 (2013).
Suparno, Sukoso, Hakim, L., Aidawati, N. Trichoderma spp. as agent of biological control in local rice diseases in Tidal swamp lands in South kalimantan, Indonesia. IOSR J. Agri. Veter. Sci. (IOSR-JAVS) 9, 01–06 (2016).
Kullnig-Gradinger, C. M., Szakacs, G. & Kubicek, C. P. Molecular identification of Trichoderma species from Russia Siberia and the Himalaya. Mycol. Res. 104, 1117–1125 (2000).
Samuels, G. J., Dodd, S. L., Gams, W., Castlebury, L. A. & Petrini, O. Trichoderma species associated with the green mould epidemic of commercially grown Agaricus bisporus. Mycologia 94, 146–170 (2002).
Hermosa, M. R. et al. Genetic diversity shown in Trichoderma biocontrol isolates. Mycol. Res. 108, 897–906 (2004).
Yadav, J., Verma, J. P. & Tiwari, K. N. Plant growth promoting activities of fungi and their effect on chickpea plant growth. Asian J. BiolSci. 4, 291–299 (2011).
Franca, D. V. C., Kupper, K. C., Magri, M. M. R. Trichoderma spp. isolates with potential of phosphate solubilization and growth promotion in cherry tomato. Pesqui. Agropecu. Trop. 47, 4 (2017).
Resende, M. P. et al. Phosphate solubilization and phytohormone production by endophytic and rhizosphere Trichoderma isolates of Guanandi (Calophyllum brasiliense Cambess). Afr. J. Micr. Res. 8, 2616–2623 (2014).
Ng, L. C., Ngadin, A., Azhari, M. & Zahari, N. A. Potential of Trichoderma spp. as biological control agents against bakanae pathogen (Fusarium fujikuroi) in Rice. As. J. Plant Pathol. 9, 46–58 (2015).
Pozo, M. J. & Azcón-Aguilar, C. Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398 (2007).
Jamalizadeh, M., Etebarian, H. R., Aminian, H. & Alizadeh, A. A review of mechanisms of action of biological control organisms against post-harvest fruit spoilage. Bull. OEPP/EPPO Bull. 41, 65–71 (2011).
Ojha, S. & Chatterjee, N. C. Mycoparasitism of Trichoderma spp. in biocontrol of fusarial wilt of tomato. Arch. Phytopathol. Plant Prot. 44, 771–782 (2011)
Qualhato, F. T. et al. Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: evaluation of antagonism and hydrolytic enzyme production. Biotech. Lett. 35, 1461–1468 (2013).
Gajera, H. P. & Vakharia, D. N. Production of lytic enzymes by Trichoderma isolates during in vitro antagonism with Aspergillus niger, the causal agent of collar rot of peanut. Braz. J. Microbiol. 43, 43–52 (2012).
El-Katatny, M. H., Somitsch, W., Robra, K. H., El-Katatny, M. S. & Gubitz, G. M. Production of chitinase and β-1, 3-glucanase by Trichoderma harzianum for control of the phytopathogenic fungus Sclerotium rolfsii. Food Technol. Biotechnol. 38, 173–180 (2000).
Maria Teresa, F. B., Adilson, L. L. & Ulhoa, C. J. Purification and characterization of an exo-Î2-1, 3-glucanase produced by Trichoderma asperellum. FEMS Microbiol. Lett. 219, 81–85 (2003).
De Marco, J. L. & Felix, C. R. Characterization of a protease produced by a Trichoderma harzianum isolate which controls cocoa plant witches’ broom disease. BMC Biochem. 3, 3 (2002).
Warin I, et al. Bioactive compound of antifungal metabolite from Trichoderma harzianum mutant strain for the control of Anthracnose of Chili (Capsicum annuum L.). Philip. Agri. Sci. 92, 392–397 (2009).
Chen, Y. T. Effects of antimicrobial peptides from Trichoderma virens FT-333 and Bacillus amyloliquefaciens BACY-1 on defense responses in rice Master Thesis (National Chiayi University, 2012).
Jeerapong, C., Phupong, W., Bangrak, P., Intana, W. & Tuchinda, P. Trichoharzianol, a new antifungal from Trichoderma harzianum F031. J. Agric. Food Chem. 63, 14 (2015).
Niranjan Raj, S., Shetty, N. P. & Shetty, H. S. Synergistic effects of Trichoshield on enhancement of growth and resistance to downy mildew in pearl millet. BioControl 50, 493–509 (2005).
Siddaiah, C. N. et al. Elicitation of resistance and associated defense responses in Trichoderma hamatum induced protection against pearl millet downy mildew pathogen. Sci. Rep. 21, 43991 (2017).
Lavanya, S. N., Niranjan-Raj, S., Nayaka, S. C. & Amruthesh, K. N. Systemic protection against pearl millet downy mildew disease induced by cell wall glucan elicitors from Trichoderma hamatum UOM 13. J. Plant Prot. Res. 57, 298–308 (2017).
O’Kennedy, M. M. et al. Expression of a β-1,3-glucanase from a biocontrol fungus in transgenic pearl millet. S. Afr. J. Bot. 77(2), 335–345 (2011).
Nandini, B. et al. Elicitation of novel trichogenic-lipid nanoemulsion signaling resistance against pearl millet downy mildew disease. Biomolecules 10, 25 (2020).
La Spada, F., Stracquadanio, C., Riolo, M., Pane, A., & Cacciola, S. O. Trichoderma counteracts the challenge of Phytophthora nicotianae infections on tomato by modulating plant defense mechanisms and the expression of crinkler, necrosis-inducing Phytophthora Protein 1, and cellulose-binding elicitor lectin pathogenic effectors. Front. Plant Sci. 11, 583539.
Peng, K. C. et al. Expression of L-amino acid oxidase of Trichoderma harzianum in tobacco confers resistance to Sclerotinia sclerotiorum and Botrytis cinerea. Plant Sci. 303, 110772 (2021).
Kamoun, S. Molecular genetics of pathogenic oomycets. Eukaryot. Cell 2, 191–199 (2003).
Martin, J. P. Use of acid, rose Bengal and streptomycin in the plate method for estimating soil fungi. Soil Sci. 69, 215–232 (1950).
Rifai, M. A. A revision of the genus Trichoderma. Mycol. Pap. 116, 1–116 (1969).
Leahy, J. G. & Colwell, R. R. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54, 305–315 (1990).
Saghai-Maroof, M. A., Soliman, K., Jorgensen, R. A. & Allard, R. W. Ribosomal DNA spacer length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proceed. Nat. Acad. Sci. 81, 8014–8018 (1984).
White, T. J., Bruns, T. D., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal genes form phylogenetics. In PCR protocols (eds Innis, M. A. et al.) 315–322 (Academic Press, 1990).
Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 (1993).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaudoise Sci. Nat. 44, 223–270 (1908).
Rohlf, F. J. NTSYS-pc numerical taxonomy and multivariate analysis system. Version 5.1. Exeter Publishing Ltd, Setauket (2001).
Botstein, D., White, R. L., Skolnick, M. & Davis, R. W. Construction of a genetic-linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331 (1980).
Powell, W. et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) marker for germplasm analysis. Mol. Breed. 2, 225–238 (1996).
Bric, J. M., Bostock, R. M. & Silverstone, S. E. Rapid in situ assay for indole acetic-acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538 (1991).
Zhang, F. et al. Putative Trichoderma harzianum mutant promotes cucumber growth by enhanced production of indole acetic acid and plant colonization. Pl. soil 368, 433–444 (2013).
Schwyn, B. & Neilands, J. B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56 (1987).
Zeilinger, S. et al. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fung. Genet. Biol. 26, 131–140 (1999).
Sadasivam, S. & Manickam, A. Biochemical methods for agricultural science, 8–9 Wiley Estern Ltd. New Delhi, India pp 246 (1992).
Lee, Y. P. & Takahashi, T. An improved colorimetric determination of amino acids with the use of ninhydrin. Anal. Biochem. 14, 71–77 (1966).
Bradford, M. M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
International Seed Testing Association (ISTA). International Seed Testing Association. Proceedings of the international seed testing association. international rules of seed testing. Seed Sci. Tech. 15, 1–9 (2005).
Abdul Baki, A. A. & Anderson, J. D. Vigor determination in soybean seed by multiple criteria. Crop Sci. 13, 630–633 (1973).
Safeeulla, K. M. Biology and control of the downy mildew of pearl millet, sorghum and finger millet (Wesley Press, 1976).
Singh, S. D. & Gopinath, R. A seedling inoculation technique for detecting downy mildew resistance in pearl millet. Plant Dis. 69, 582–584 (1985).
Williams, R. J., Singh, S. D. & Pawar, M. N. An improved field screening for downy mildew resistance in pearl millet. Plant Dis. 65, 239–241 (1981).
Acknowledgements
The authors are thankful to SERB (DST), New Delhi for providing research funding through Fast-track for Major research project for Young Scientists. The authors are also thankful to the Indian Council of Agricultural Research (ICAR), Government of India, New Delhi, for providing field facilities (Downy mildew sick plot) at the Department of studies in Biotechnology, University of Mysore and Institution of Excellence (IOE) Project Authorities University of Mysore.
Author information
Authors and Affiliations
Contributions
B.N., carried out the experimental work and wrote the manuscript. G N., P.H., R. K. S., and H.S.P., designed the experiment, analyzed data, and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nandini, B., Puttaswamy, H., Saini, R.K. et al. Trichovariability in rhizosphere soil samples and their biocontrol potential against downy mildew pathogen in pearl millet. Sci Rep 11, 9517 (2021). https://doi.org/10.1038/s41598-021-89061-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89061-2
- Springer Nature Limited
This article is cited by
-
With a little help from my friends: inoculation with Bacillus amyloliquefaciens and Trichoderma asperellum alleviates drought and salt stress in soybean
Theoretical and Experimental Plant Physiology (2023)
-
Biological control of groundnut stem rot using native isolates of Trichoderma harzianum and T. aggressivum f. aggressivum
Journal of Plant Pathology (2022)