Abstract
Meta-analysis can be applied to study the effectiveness of the summary estimates for experimental papers, producing objective and unbiased results. We investigated the effects of phosphoinositide-3-kinase (PI3K) on the inflammatory profile in allergic mouse models, which are currently under development in signal transduction materials. PubMed, EMBASE and Web of Science databases were searched for relevant literature using the search terms “ PI3K inhibitor” and “allergy” or “asthma”. Cochrane Review Manager and R were used for handling continuous variables. The primary outcomes of the inflammatory profile were divided into cell counts and inflammatory cytokines. We used a random effects model to draw a forest plot. Through the database search and subsequent selection, 17 articles were identified. Regarding the cell counts, both the PI3K pan-inhibitors and PI3K-δ inhibitors effectively reduced the total cell counts, eosinophils, neutrophils and lymphocytes. In contrast to PI3K-δ inhibitors, PI3K pan-inhibitors effectively reduced macrophages. Regarding the inflammatory cytokines, PI3K pan-inhibitors and PI3K-δ inhibitors effectively reduced total IgE, IL-4, IL-5, IL-13, TNF-α, IL-1β, VEGF and had no effect on IL-6. Compared to the PI3K pan-inhibitors, which block all pathways, selective PI3K-δ inhibitors are expected to be relatively less toxic. Regarding the efficacy, PI3K-δ inhibitors have at least the same or better efficacy than PI3K pan-inhibitors in effector cells and inflammatory mediators.
Similar content being viewed by others
Introduction
Meta-analysis is a method of collecting and organizing data based on existing documents and deriving the most objective and unbiased results in a transparent way. As is well known, existing meta-analyses in Cochrane Reviews have used interventional meta-analytic methods. Interventional meta-analysis is mainly used to ensure the internal legitimacy of study results. Meta-analysis can be applied to determine the effectiveness of the summary estimates for the experimental papers included in the analysis, and thus the objective basis of the study is secured1. Clinical trials in the area of pathophysiology are particularly limited, while animal studies can investigate the pathophysiology more extensively. These animal studies are somewhat time consuming, difficult, less reproducible, and costly. Applying meta-analysis in animal studies dealing with pathophysiology can save costs and synthesize the results of each discrete study, resulting in very meaningful final results.
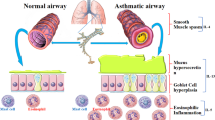
Class I phosphoinositide 3-kinases (PI3Ks), lipid signalling kinases acting adjacent to cell membrane, have emerged as a promising therapeutic target for allergic inflammatory disorders because they are implicated in a broad aspect of cellular pathophysiology, and that interfering this signalling allows the development of potent anti-inflammatory agent effective against broad spectrum of disease severity2. PI3Ks are divided into four subtypes (α, β, γ, δ). Whereas PI3K-α and -β isoforms are present everywhere, PI3K-δ is an isoform which is uniquely distributed in circulating haematogenous cells and endothelial cells, making it particularly attractive druggable target for immune/inflammatory disorders including bronchial asthma3. Since the beginning of the research, therapeutic blockade of pan-class I PI3Ks have been vastly studied in oncology such as solid cancer and lymphoma, and researches have been also conducted in the field of allergy4. As for PI3K-δ isoform, it is also increasingly recognized as a critical mediator of severe allergic inflammation in upper respiratory tract as well as lower airways2,5. However, at the same time, there are concerns on the blockade of various isoforms of PI3Ks for therapeutic purposes due to its potential to disturb protective immune and inflammatory responses against diverse pathogens. In this regard, comparative analysis of the therapeutic effects of PI3Ks pan-inhibition and PI3K-δ selective blockade on different aspects of immune/inflammatory process using validated method is important for a novel drug development for the disease.
Based on this knowledge, through a meta-analysis on the pre-existing preclinical data, we investigate the therapeutic effects of both PI3K pan-inhibitor and PI3K-δ selective inhibitor on features of allergic lung inflammation including inflammatory cell infiltrations into lungs and levels of various pro-inflammatory cytokines/chemokines in several murine models of allergic lung inflammation.
Methods
Search strategy
This study was the subject of IRB exemption from Chonbuk National University Hospital, and followed the PRISMA guidelines6. PubMed, EMBASE, Web of Science, Google Scholar, and Cochrane Library databases were included in the literature search. The search term in Medline was: (PI3K inhibitor OR idelalisib) AND (asthma OR allergic OR allergy). All the PI3K pan-inhibitor and PI3K-δ selective inhibitor found in our search criteria. Similar search terms were used for the four other databases. Two authors independently performed the literature search and the published studies identified in the search results included data up to July 2019.
Selection of individual studies
The inclusion criteria for studies were that they: (1) discussed the PI3K signalling pathway; (2) discussed allergies or asthma; (3) were in English; (4) contained meaningful data (cell counts and inflammatory cytokines); (5) included an allergy or asthma mouse model.
Exclusion criteria for studies were that they: (1) did not mention PI3K inhibitor; (2) discussed leukaemia or other malignancies; (3) did not contain appropriate data; (4) were review articles; (5) discussed organs other than those related to asthma; (6) did not address the inflammatory profile; (7) included animal models other than allergy or asthma; or (8) dealt with PI3K-δ and γ dual inhibitor.
We divided the PI3K inhibitors into two groups as follows: (1) drugs that inhibited all PI3K isoforms (α, β, γ, and δ) were regarded as PI3K pan-inhibitors; (2) drugs that specifically inhibited PI3K-δ were termed PI3K-δ inhibitors. Individual studies were selected only when used in an allergic lung inflammatory mouse model.
Data extraction
Three authors independently screened the study titles and abstracts (JSK, JSJ, SHK). The following information was extracted from each study: author names, year of publication, allergy model, PI3K drug name, dose used, inflammatory profile, duration of study, population used. When any disagreement occurred, the other authors acted as arbitrators.
Statistical analysis
Cochrane Review Manager and R were used for statistical analysis. First, we found the mean and SD values for the continuous variables in the inflammatory profile in the original text. If the data were not presented numerically in the original paper, the mean and SD values were calculated from a graph using the Graph Data Extractor7. The primary outcomes of the inflammatory profile were as follows: total cell counts, eosinophils, neutrophils, macrophages, lymphocytes, total IgE, IL-4, IL-5, IL-13, eotaxin, IFN-γ, IL-6, TNF-α, TGF-β, IL-1β, VEGF. Data were analyzed as continuous variables. Standard mean differences (SMD with 95%CI), Cohen’s d and Hedge’s g were calculated from the mean and SD values8,9. Standard error (SE) was calculated by the inverse variance method. The I2 value was used to assess inter-study heterogeneity. We used a random effects model to draw a forest plot, which we considered to be a suitable model for animal studies where there may be many heterogeneities1.
Mathmatical formula in this meta-analysis
We followed standard meta-analysis techniques1,9,10. The effect size from each individual independent study was obtained as outlined next.
The variables used in this study are all continuous variables, and d is the standardized mean difference between the two groups \({\bar{X}}_{1}\) and \({\bar{X}}_{2}\).
In the numerator, \({\bar{X}}_{1}\) and \({\bar{X}}_{2}\) are the sample means in the two groups. In the denominator, Swithin is the within-groups standard deviation, pooled across groups, where n1 and n2 are the sample sizes in the two groups, and S1 and S2 are the standard deviations in the two groups.
Variations of d are as follows.
The total variance for a study \(V{Y}_{i}^{\ast }\) is the sum of the within-study variance (VYi) and the between-studies variance (T2). This method of estimating the variance between studies is the most popular, and is known as the DerSimonian and Laird method9.
To combine the studies, we used a random-effects model in which studies are weighted by the sum of the true variation among studies and the sampling variation within studies. The effect sizes were combined across studies to give a weighted mean effect size across K studies:
where θ is the effect size of each gene in each study, and wi is the corresponding weight for that study.
Results
Search results
Using the search terms as discussed earlier, a total of 771 articles were identified: 184 articles in Medline, 241 articles in Web of Science, 151 articles in EMBASE, 192 articles in Google Scholar, and 3 articles in Cochrane Library. From these, the title and abstract were checked, and 540 articles were excluded as irrelevant studies, leaving 231 articles. Of these, 176 articles were excluded as duplicate papers, leaving 55 full length articles. Of these, a total of 38 papers were excluded: 3 papers on leukaemia, 12 papers without data, 14 papers with insufficient data, and 4 papers with duplicate data. Thus, a total of 17 papers were left for qualitative and quantitative evaluation (Tables 1 and 2)11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27.
Eleven of these studies were on PI3K pan-inhibitors (Table 1), and six were on PI3K-δ inhibitors (Table 2). Primary outcomes were cell counts and inflammatory profile. We evaluated the effects of PI3K pan-inhibitors and PI3K-δ inhibitors as subgroups. The PRISMA diagram with the main selection process and reasons for exclusion is shown in Fig. 1.
Effector cell counts
Total cell counts
The efficacy of the PI3K pan-inhibitors on total cell counts was SMD: −3.56 [95% CI: −5.55 to −1.57] with substantial heterogeneity (I2 = 87%) in a total of 9 articles (Fig. 2A). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on total cell counts was SMD: −5.56 [95% CI: −7.74 to −3.38] with moderate heterogeneity (I2 = 71%) in a total of 5 articles (Fig. 2B).
Forest plots regarding effector cell counts showing the effect of: (A) PI3K pan-inhibitors on total cell counts; (B) PI3K-δ inhibitors on total cell counts; (C) PI3K pan-inhibitors on eosinophil counts; (D) PI3K-δ inhibitors on eosinophil counts; (E) PI3K pan-inhibitors on neutrophil counts; (F) PI3K-δ inhibitors on neutrophil counts; (G) PI3K pan-inhibitors on macrophage counts; (H) PI3K-δ inhibitors on macrophage counts; (I) PI3K pan-inhibitors on lymphocyte counts; (J) PI3K-δ inhibitors on lymphocyte counts.
Eosinophils
The efficacy of the PI3K pan-inhibitors on eosinophil counts was SMD: −2.70 [95% CI: −3.53 to −1.86] with moderate heterogeneity (I2 = 62%) in a total of 11 articles (Fig. 2C). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on eosinophil counts was SMD: −8.67 [95% CI: −12.37 to −4.97] with substantial heterogeneity (I2 = 79%) in a total of 5 articles (Fig. 2D).
Neutrophils
The efficacy of the PI3K pan-inhibitors on neutrophil counts was SMD: −4.05 [95% CI: −6.17 to −1.93] with substantial heterogeneity (I2 = 89%) in a total of 10 articles (Fig. 2E). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on neutrophil counts was SMD: −3.62 [95% CI: −5.66 to −1.59] with substantial heterogeneity (I2 = 85%) in a total of 5 articles (Fig. 2F).
Macrophages
The efficacy of the PI3K pan-inhibitors on macrophage counts was SMD: −2.06 [95% CI: −3.69 to −0.43] with substantial heterogeneity (I2 = 88%) in a total of 8 articles (Fig. 2G). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on macrophage counts was SMD: 1.59 [95% CI: 0.21 to 2.96] in a total of 1 article (Fig. 2H).
Lymphocytes
The efficacy of the PI3K pan-inhibitors on lymphocyte counts was SMD: −2.56 [95% CI: −4.56 to −0.56] with substantial heterogeneity (I2 = 89%) in a total of 7 articles (Fig. 2I). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on lymphocyte counts was SMD: −3.35 [95% CI: −4.38 to −2.33] with low heterogeneity (I2 = 37%) in a total of 5 articles (Fig. 2J).
Inflammatory cytokines and chemokines
Total IgE
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on total IgE was SMD: −5.06 [95% CI: −6.80 to −3.32] with moderate heterogeneity (I2 = 35%) in a total of 3 articles (Fig. 3A). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on total IgE was SMD: −4.37 [95% CI: −6.18 to −2.55] with no heterogeneity (I2 = 0%) in a total of 2 articles (Fig. 3B).
Forest plots regarding inflammatory cytokines and chemokines showing the effect of: (A) PI3K pan-inhibitors on total IgE; (B) PI3K-δ inhibitors on total IgE; (C) PI3K pan-inhibitors on IL-4; (D) PI3K-δ inhibitors on IL-4; (E) PI3K pan-inhibitors on IL-5; (F) PI3K-δ inhibitors on IL-5; (G) PI3K pan-inhibitors on IL-13; (H) PI3K-δ inhibitors on IL-13; (I) PI3K pan-inhibitors on eotaxin; (J) PI3K-δ inhibitors on eotaxin; (K) PI3K pan-inhibitors on IFN-γ; (L) PI3K pan-inhibitors on TGF-β; (M) PI3K pan-inhibitors on IL-6; (N) PI3K-δ inhibitors on IL-6; (O) PI3K pan-inhibitors on TNF-α; (P) PI3K-δ inhibitors on TNF-α; (Q) PI3K pan-inhibitors on IL-1β; (R) PI3K-δ inhibitors on IL-1β; (S) PI3K pan-inhibitors on VEGF; (T) PI3K-δ inhibitors on VEGF.
IL-4
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on IL-4 was SMD: −3.20 [95% CI: −5.44 to −0.95] with substantial heterogeneity (I2 = 89%) in a total of 7 articles (Fig. 3C). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on IL-4 was SMD: −1.95 [95% CI: −3.09 to −0.80] with moderate heterogeneity (I2 = 52%) in a total of 3 articles (Fig. 3D).
IL-5
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on IL-5 was SMD: −3.51 [95% CI: −5.63 to −1.39] with substantial heterogeneity (I2 = 88%) in a total of 8 articles (Fig. 3E). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on IL-5 was SMD: −2.98 [95% CI: −3.96 to −2.00] with low heterogeneity (I2 = 7%) in a total of 3 articles (Fig. 3F).
IL-13
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on IL-13 was SMD: −2.99 [95% CI: −5.71 to −0.27] with substantial heterogeneity (I2 = 89%) in a total of 6 articles (Fig. 3G). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on IL-13 was SMD: −2.75 [95% CI: −3.61 to −1.88] with no heterogeneity (I2 = 0%) in a total of 3 articles (Fig. 3H).
Eotaxin
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on eotaxin was SMD: −4.96 [95% CI: −8.22 to −1.70] with moderate heterogeneity (I2 = 68%) in a total of 3 articles (Fig. 3I). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on eotaxin was SMD: −3.50 [95% CI: −11.01 to 4.00] with substantial heterogeneity (I2 = 93%) in a total of 2 articles (Fig. 3J).
IFN-γ
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on IFN-γ was SMD: −0.20 [95% CI: −5.18 to 4.79] with substantial heterogeneity (I2 = 93%) in a total of 3 articles (Fig. 3K).
TGF-β
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on TGF-β was SMD: −3.23 [95% CI: −5.43 to −1.02] with moderate heterogeneity (I2 = 47%) in a total of 2 articles (Fig. 3L).
IL-6
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on IL-6 was SMD: −4.26 [95% CI: −9.83 to 1.32] with substantial heterogeneity (I2 = 83%) in a total of 2 articles (Fig. 3M). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on IL-6 was SMD: −5.34 [95% CI: −10.75 to 0.06] in one article (Fig. 3N).
TNF-α
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on TNF-α was SMD: −4.21 [95% CI: −6.02 to −2.39] with low heterogeneity (I2 = 26%) in a total of 3 articles (Fig. 3O). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on TNF-α was SMD: −5.66 [95% CI: −8.31 to −3.00] with no heterogeneity (I2 = 0%) in a total of 2 articles (Fig. 3P).
IL-1β
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on IL-1β was SMD: −6.24 [95% CI: −10.86 to −1.61] with substantial heterogeneity (I2 = 82%) in a total of 3 articles (Fig. 3Q). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on IL-1β was SMD: −1.45 [95% CI: −2.78 to −0.11] in one article (Fig. 3R).
VEGF
Among the inflammatory profiles, the efficacy of the PI3K pan-inhibitors on VEGF was SMD: −5.18 [95% CI: −7.86 to −2.49] with low heterogeneity (I2 = 23%) in a total of 2 articles (Fig. 3S). As a subgroup analysis, the efficacy of the PI3K-δ inhibitors on VEGF was SMD: −6.83 [95% CI: −9.73 to −3.94] in one article (Fig. 3T).
Summary
Regarding the cell counts, both the PI3K pan-inhibitors and PI3K-δ inhibitors effectively reduced the total cell counts, eosinophils, neutrophils, and lymphocytes. In contrast to PI3K-δ inhibitors, PI3K pan-inhibitors effectively reduced macrophages.
Regarding the inflammatory cytokines, PI3K pan-inhibitors and PI3K-δ inhibitors effectively reduced total IgE, IL-4, IL-5, IL-13, TNF-α, IL-1β, and VEGF, and had no effect on IL-6. PI3K pan-inhibitors reduced TGF-β and eotaxin, but had no effect on IFN-γ. PI3K-δ inhibitors had no effect on eotaxin.
Discussion
Before the 1990s, narrative reviews took the role of combining data from multiple studies. A narrative review is a form in which an expert in one field draws conclusions by synthesizing previous studies on a subject. This form has inherent problems of subjectivity due to the lack of transparency. To solve these problems, since the 1990s, there has been a move to apply a new form of systematic review and meta-analysis in various disciplines such as medicine, pharmacy, education, psychology, sociology and ecology9. Unlike the narrative review, in which a researcher arbitrarily assigns a weight to each individual study, a meta-analysis secures objectivity by weighting each individual study according to mathematical criteria specially designed in advance. The statistical analysis technique used in the meta-analysis is a transparent, objective, and repeatable framework. This meta-analysis is used to support evidence-based policy and to obtain evidence on the effectiveness of any intervention for a variety of reasons.
In the current study, we present the results of a meta-analytic method which was used to investigate therapeutic effects of PI3K pan-inhibitors/PI3K-δ inhibitors in various experimental murine models of allergic lung inflammation.
Allergic inflammation is orchestrated by complex interplays between diverse cellular components of innate and adaptive immunity in the background of immense pro-inflammatory milieus, in which a wide array of cytokines and chemokines are closely implicated. Bronchial asthma is the hallmark of the allergic inflammatory disorder in the lower respiratory tract28. Unfortunately, there is no curable agent currently available for the treatment of allergy and asthma, although numerous therapeutic options have been developed. Some of them are not sufficiently effective for the refractory disease (i.e. corticosteroid resistance in patients having severe asthma) and others are efficacious for specific subset of severe asthmatic patients, however, they are high-priced that precludes widespread use of those agents (e.g. biologicals that target type 2 cytokines such as IL-4, 5, 13)29,30. In this regard, class I PI3Ks have been gaining much attention as promising therapeutic target for allergic disorders because their widespread involvement in controlling nearly all aspects of cellular events, including growth, proliferation, metabolism, motility, and survival2,4. Initial studies on the PI3K pathway were mainly driven by cancer biologists31. Furthermore, with the increasing knowledge on this pathway, PI3K-targeted therapies using PI3K pan-inhibitor have revealed that the PI3K pathway are closely implicated in a broad spectrum of immune/inflammatory diseases including allergy and bronchial asthma21.
Meanwhile, crucial involvement of certain isoforms of PI3Ks in normal physiologic process (e.g. genetic knockdown of PI3K-α and -β isoforms leads to embryonic lethality32,33) raised concerns on the use of PI3K pan-inhibitors for therapeutic purpose due to its non-selectivity enough to cause systemic adverse effects. In this context, therapeutic blockade of specific isoform of PI3Ks such as PI3K-δ has been intensively studied particularly in allergic inflammation26,34, given its preferential expression in hematogenous immune/inflammatory cells such as leukocytes. This approach may reduce potential harmful effects mediated through interfering the normal physiologic and protective inflammatory responses against invading microorganisms. Nonetheless, limited information exists regarding comparative analysis on the therapeutic effects of PI3K pan-inhibition and δ isoform selective inhibition in the treatment of allergic lung inflammation, partly because many of these agents are in the early stage of development, so that their clinical efficacies in real practice are not characterized thoroughly yet2,5. Thus, through evaluating their therapeutic effects on multifaceted process of allergic inflammation in pre-clinical experimental models, we can get much information on their comparative efficacies, and thus this may facilitate the development of a novel PI3K-targeted therapy. Particularly, considering the cost and time of each animal experiment, we were interested in new research methods that could improve the integration of results from previous studies. In this regard, a systematic review and meta-analysis of PI3K pan-inhibitors and PI3K-δ inhibitors in animal studies was thought to be an attractive and novel beneficial approach.
As for diverse cellular components of allergic lung inflammation, our results showed that PI3K pan-inhibitors and PI3K-δ inhibitors effectively reduced total cell counts, neutrophils, lymphocytes, eosinophils, and macrophages. Eosinophils are leukocytes that have multiple functions in the host defence and are also involved in immune regulation. Neutrophils are also major key factors in the epithelial barrier in allergic disease and are associated with the severity of allergic asthma35,36. Eosinophils are also involved in the production of inflammatory mediators while releasing toxic granule proteins such as eosinophil cationic protein, eosinophil peroxidase, and eosinophil-derived neurotoxin37. The emergence of eosinophils from the bone marrow is a particularly important process in the allergic inflammatory reaction, and is regulated by IL-514. PI3K pan-inhibitors are known to inhibit this process. Although the PI3K pan-inhibitors have been shown to effectively lower neutrophils and eosinophils, it has been shown that PI3K-δ selective inhibitors also effectively lower both effector cells.
In particular, IL-4, -5 and -13 are produced in Th2 cells and are associated with allergic hyperresponsiveness and are deeply involved in airway inflammation through eosinophil activation38. IL-4, -5, and -13 are known to play crucial roles in the production, migration, survival, and activation of eosinophils. Our results have shown that both PI3K pan-inhibitors and PI3K-δ selective inhibitors effectively lowered IL-4, IL-5, and IL-13, and also effectively lowered the number of eosinophils, the major effector cell.
Classically, IL-1β and TNF-α are necessary for the enhancement of eosinophil migration, activation, and survival, and are necessary for the enhancement of allergic hyperresponsiveness18. In fact, IL-1β has been shown to be involved in inflammatory diseases. The inflammasome promotes the maturation and secretion of the pro-inflammatory cytokine IL-1β. TNF-α has similar biologic activity, and both are involved in innate immunity39,40,41. Our study has shown that PI3K pan-inhibitors effectively inhibited both IL-1β and TNF-α, and PI3K-δ selective inhibitors can also inhibit these two important cytokines.
VEGF is a vascular endothelial growth factor that is involved in the formation of blood vessels around the bronchus, and is involved in airway oedema and narrowing, and eventually participates in airway vascular remodelling42. In patients with acute asthma, it has been found that VEGF is elevated43. In addition, the level of VEGF in asthma patients is highly correlated with disease activity and has been found to be inversely proportional to the diameter of the airway44. In our study, both PI3K pan-inhibitors and PI3K-δ inhibitors effectively lowered VEGF.
TGF-β is involved in airway remodelling and fibrosis, and is involved in direct smooth muscle contraction and obstruction of the airways45. TGF-β is known to be an important factor involved in tissue fibrosis in asthma14. PI3K pan-inhibitors effectively lowered TGF-β, however, there was no data with regard to PI3K-δ inhibitors.
IL-6 is a multifunctional cytokine whose function is to regulate immune response, hematopoiesis, and inflammation39. It plays an important role in differentiation of naive T cells into Th17 lymphocytes; however, both PI3K pan-inhibitors and PI3K-δ inhibitors had no effect on IL-6.
In conclusion, selective PI3K-δ inhibitors have at least the same or better efficacy compared with PI3K pan-inhibitors against effector cells and inflammatory mediators. Since the single pathway inhibitors (PI3K-δ inhibitors) can be as effective as PI3K pan-inhibitors, it is recommended to apply them to allergies and asthma. Importantly, we have found that most of the major inflammatory actions associated with PI3K signalling, known as the critical pathway, are mediated by the delta isotype pathway. Our results will be helpful in future clinical trials studying the PI3K-δ signalling pathway.
References
Vesterinen, H. M. et al. Meta-analysis of data from animal studies: a practical guide. J Neurosci Methods 221, 92–102, https://doi.org/10.1016/j.jneumeth.2013.09.010 (2014).
Jeong, J. S., Kim, J. S., Kim, S. R. & Lee, Y. C. Defining Bronchial Asthma with Phosphoinositide 3-Kinase Delta Activation: Towards Endotype-Driven Management. Int J Mol Sci 20, https://doi.org/10.3390/ijms20143525 (2019).
Vanhaesebroeck, B., Guillermet-Guibert, J., Graupera, M. & Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 11, 329–341, https://doi.org/10.1038/nrm2882 (2010).
Goncalves, M. D., Hopkins, B. D. & Cantley, L. C. Phosphatidylinositol 3-Kinase, Growth Disorders, and Cancer. N Engl J Med 379, 2052–2062, https://doi.org/10.1056/NEJMra1704560 (2018).
Horak, F. et al. Randomized phase 1 study of the phosphatidylinositol 3-kinase delta inhibitor idelalisib in patients with allergic rhinitis. J Allergy Clin Immunol 137, 1733–1741, https://doi.org/10.1016/j.jaci.2015.12.1313 (2016).
Liberati, A. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339, b2700, https://doi.org/10.1136/bmj.b2700 (2009).
Kim, J. S., Kwon, S. H., Lee, S. E., Lee, E. J. & Lee, M. H. Effect of single-dose intravenous dexamethasone on subjective voice quality after thyroidectomy: A meta-analysis. Medicine (Baltimore) 97, e11832, https://doi.org/10.1097/MD.0000000000011832 (2018).
Kim, J. S. & Kwon, S. H. Recurrence of sinonasal inverted papilloma following surgical approach: A meta-analysis. Laryngoscope 127, 52–58, https://doi.org/10.1002/lary.26222 (2017).
Borenstein, M., Hedges, L. V., Higgins, J. P. T. & Rothstein, H. R. Introduction to meta-analysis (Wiley, 2009).
Rest, J. S., Wilkins, O., Yuan, W., Purugganan, M. D. & Gurevitch, J. Meta-analysis and meta-regression of transcriptomic responses to water stress in Arabidopsis. Plant J 85, 548–560, https://doi.org/10.1111/tpj.13124 (2016).
Campa, C. C. et al. Inhalation of the prodrug PI3K inhibitor CL27c improves lung function in asthma and fibrosis. Nat Commun 9, 5232, https://doi.org/10.1038/s41467-018-07698-6 (2018).
Wagh, A. D. et al. Investigation into the Role of PI3K and JAK3 Kinase Inhibitors in Murine Models of Asthma. Front Pharmacol 8, 82, https://doi.org/10.3389/fphar.2017.00082 (2017).
Huang, P. et al. Comprehensive attenuation of IL-25-induced airway hyperresponsiveness, inflammation and remodelling by the PI3K inhibitor LY294002. Respirology 22, 78–85, https://doi.org/10.1111/resp.12880 (2017).
Oikawa, Y., Sasaki, N., Niisato, M., Nakamura, Y. & Yamauchi, K. A phosphatidylinositol 3-kinase inhibitor strongly suppressed pulmonary vascular remodeling of allergic vasculitis in a murine model. Exp Lung Res 42, 111–120, https://doi.org/10.3109/01902148.2016.1157226 (2016).
Saw, S. & Arora, N. PI3K and ERK1/2 kinase inhibition potentiate protease inhibitor to attenuate allergen induced Th2 immune response in mouse. Eur J Pharmacol 776, 176–184, https://doi.org/10.1016/j.ejphar.2016.02.050 (2016).
Yao, L. et al. Phosphatidylinositol 3-Kinase Mediates beta-Catenin Dysfunction of Airway Epithelium in a Toluene Diisocyanate-Induced Murine Asthma Model. Toxicol Sci 147, 168–177, https://doi.org/10.1093/toxsci/kfv120 (2015).
Liang, J. et al. Phosphatidylinositol 3-kinases pathway mediates lung caspase-1 activation and high mobility group box 1 production in a toluene-diisocyanate induced murine asthma model. Toxicol Lett 236, 25–33, https://doi.org/10.1016/j.toxlet.2015.04.011 (2015).
Choi, Y. H., Jin, G. Y., Li, L. C. & Yan, G. H. Inhibition of protein kinase C delta attenuates allergic airway inflammation through suppression of PI3K/Akt/mTOR/HIF-1 alpha/VEGF pathway. PLoS One 8, e81773, https://doi.org/10.1371/journal.pone.0081773 (2013).
Xia, X. et al. Phosphatidylinositol 3-kinase inhibitor suppresses inducible nitric oxide synthase expression in bronchiole epithelial cells in asthmatic rats. Mol Cell Biochem 359, 293–299, https://doi.org/10.1007/s11010-011-1023-y (2012).
Duan, W., Aguinaldo Datiles, A. M., Leung, B. P., Vlahos, C. J. & Wong, W. S. An anti-inflammatory role for a phosphoinositide 3-kinase inhibitor LY294002 in a mouse asthma model. Int Immunopharmacol 5, 495–502, https://doi.org/10.1016/j.intimp.2004.10.015 (2005).
Kwak, Y. G. et al. Involvement of PTEN in airway hyperresponsiveness and inflammation in bronchial asthma. J Clin Invest 111, 1083–1092, https://doi.org/10.1172/JCI16440 (2003).
Lee, K. S. et al. Phosphoinositide 3-kinase-delta regulates fungus-induced allergic lung inflammation through endoplasmic reticulum stress. Thorax 71, 52–63, https://doi.org/10.1136/thoraxjnl-2015-207096 (2016).
Collmann, E. et al. Transient targeting of phosphoinositide 3-kinase acts as a roadblock in mast cells’ route to allergy. J Allergy Clin Immunol 132, 959–968, https://doi.org/10.1016/j.jaci.2013.03.008 (2013).
Kang, B. N. et al. The p110delta subunit of PI3K regulates bone marrow-derived eosinophil trafficking and airway eosinophilia in allergen-challenged mice. Am J Physiol Lung Cell Mol Physiol 302, L1179–1191, https://doi.org/10.1152/ajplung.00005.2012 (2012).
Lee, K. S. et al. Phosphoinositide 3-kinase-delta inhibitor reduces vascular permeability in a murine model of asthma. J Allergy Clin Immunol 118, 403–409, https://doi.org/10.1016/j.jaci.2006.04.041 (2006).
Lee, K. S., Lee, H. K., Hayflick, J. S., Lee, Y. C. & Puri, K. D. Inhibition of phosphoinositide 3-kinase delta attenuates allergic airway inflammation and hyperresponsiveness in murine asthma model. FASEB J 20, 455–465, https://doi.org/10.1096/fj.05-5045com (2006).
Park, S. J. et al. Phosphoinositide 3-kinase delta inhibitor suppresses interleukin-17 expression in a murine asthma model. Eur Respir J 36, 1448–1459, https://doi.org/10.1183/09031936.00106609 (2010).
Lambrecht, B. N., Hammad, H. & Fahy, J. V. The Cytokines of Asthma. Immunity 50, 975–991, https://doi.org/10.1016/j.immuni.2019.03.018 (2019).
Carr, T. F., Zeki, A. A. & Kraft, M. Eosinophilic and Noneosinophilic Asthma. Am J Respir Crit Care Med 197, 22–37, https://doi.org/10.1164/rccm.201611-2232PP (2018).
Jeong, J. S., Kim, S. R., Cho, S. H. & Lee, Y. C. A Novel Insight on Endotyping Heterogeneous Severe Asthma Based on Endoplasmic Reticulum Stress: Beyond the “Type 2/Non-Type 2 Dichotomy”. Int J Mol Sci 20, https://doi.org/10.3390/ijms20030713 (2019).
Liu, P., Cheng, H., Roberts, T. M. & Zhao, J. J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8, 627–644, https://doi.org/10.1038/nrd2926 (2009).
Bi, L., Okabe, I., Bernard, D. J., Wynshaw-Boris, A. & Nussbaum, R. L. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem 274, 10963–10968, https://doi.org/10.1074/jbc.274.16.10963 (1999).
Bi, L., Okabe, I., Bernard, D. J. & Nussbaum, R. L. Early embryonic lethality in mice deficient in the p110beta catalytic subunit of PI 3-kinase. Mamm Genome 13, 169–172, https://doi.org/10.1007/bf02684023 (2002).
Ali, K. et al. Essential role for the p110delta phosphoinositide 3-kinase in the allergic response. Nature 431, 1007–1011, https://doi.org/10.1038/nature02991 (2004).
Pothoven, K. L. et al. Neutrophils are a major source of the epithelial barrier disrupting cytokine oncostatin M in patients with mucosal airways disease. J Allergy Clin Immunol 139, 1966–1978 e1969, https://doi.org/10.1016/j.jaci.2016.10.039 (2017).
Vedel-Krogh, S., Fallgaard Nielsen, S., Lange, P., Vestbo, J. & Nordestgaard, B. G. Association of Blood Eosinophil and Blood Neutrophil Counts with Asthma Exacerbations in the Copenhagen General Population Study. Clin Chem 63, 823–832, https://doi.org/10.1373/clinchem.2016.267450 (2017).
Kampe, M. et al. PI3-kinase regulates eosinophil and neutrophil degranulation in patients with allergic rhinitis and allergic asthma irrespective of allergen challenge model. Inflammation 35, 230–239, https://doi.org/10.1007/s10753-011-9309-5 (2012).
Medoff, B. D., Thomas, S. Y. & Luster, A. D. T cell trafficking in allergic asthma: the ins and outs. Annu Rev Immunol 26, 205–232, https://doi.org/10.1146/annurev.immunol.26.021607.090312 (2008).
Adkinson Newton Franklin, M. E. J. Middleton’s Allergy Principles And Practice. 8th edition edn, (Elsevier, 2014).
Furuoka, M. et al. TNF-alpha Induces Caspase-1 Activation Independently of Simultaneously Induced NLRP3 in 3T3-L1 Cells. J Cell Physiol 231, 2761–2767, https://doi.org/10.1002/jcp.25385 (2016).
Satoh, T., Otsuka, A., Contassot, E. & French, L. E. The inflammasome and IL-1beta: implications for the treatment of inflammatory diseases. Immunotherapy 7, 243–254, https://doi.org/10.2217/imt.14.106 (2015).
McCullagh, A. et al. The bronchial circulation–worth a closer look: a review of the relationship between the bronchial vasculature and airway inflammation. Pediatr Pulmonol 45, 1–13, https://doi.org/10.1002/ppul.21135 (2010).
Lee, Y. C. & Lee, H. K. Vascular endothelial growth factor in patients with acute asthma. J Allergy Clin Immunol 107, 1106 (2001).
Kanazawa, H. VEGF, angiopoietin-1 and -2 in bronchial asthma: new molecular targets in airway angiogenesis and microvascular remodeling. Recent Pat Inflamm Allergy Drug Discov 1, 1–8 (2007).
Adler, K. B. D. T. Growth Factor-beta Induce Persistent Airway Obstruction after Asthma Exacerbations? Am J Respir Cell Mol Biol 58, 543–544, https://doi.org/10.1165/rcmb.2017-0339ED (2018).
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2017R1A2A1A05000747; Y.C.L.) and the fund of Biomedical Research Institute, Jeonbuk National University Hospital, and by research funds for newly appointed professors of Jeonbuk National University in 2018 (J.S.K).
Author information
Authors and Affiliations
Contributions
Acquisition of data: J.S.K., J.S.J.; analysis and interpretation of data: J.S.K., J.S.J.; Drafting the manuscript for critical intellectual content: J.S.K., J.S.J.; review and improvement of the manuscript: S.R.K., Y.C.L., S.H.K.; Conception and design of the study: J.S.K., J.S.J., Y.C.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, J.S., Jeong, J.S., Kwon, S.H. et al. Roles of PI3K pan-inhibitors and PI3K-δ inhibitors in allergic lung inflammation: a systematic review and meta-analysis. Sci Rep 10, 7608 (2020). https://doi.org/10.1038/s41598-020-64594-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-64594-0
- Springer Nature Limited
This article is cited by
-
Development of phosphoinositide 3-kinase delta (PI3Kδ) inhibitors as potential anticancer agents through the generation of ligand-based pharmacophores and biological screening
Medicinal Chemistry Research (2023)
-
Network pharmacology and molecular docking to explore the potential mechanism of urolithin A in combined allergic rhinitis and asthma syndrome
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Regulatory mechanisms of neutrophil migration from the circulation to the airspace
Cellular and Molecular Life Sciences (2021)