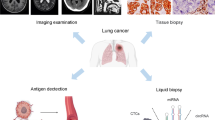
Abstract
Purpose
Liquid biopsy is a promising technological method in patient management of early-stage non-small-cell lung cancer (NSCLC). The detection platforms exhibit high efficiency and related clinical applications also emerge with high-quality performance. An overview of the current status is in need for an integrated perception on this field.
Methods
NSCLC takes up the largest proportion of lung cancer and there is a tendency for more early-stage patients in real practice. Hence, early-stage NSCLC participants occupy an important position in clinical work. Liquid biopsy, as a promising non-invasive detection method, had great potential in various aspects of the whole diagnosis-treatment procedure. We went through the landmark articles according to liquid biopsy in the field of early-stage NSCLC management and concluded the status quo of it.
Results
In this review, we summarized the improvement of the detection technologies regarding the most widely studied biomarkers and elucidated the current clinical applications of liquid biopsy in early detection, prognostic performance assessment, and predictive value respectively, in early-stage NSCLC patients.
Conclusion
Liquid biopsy has achieved favorable outcomes in different aspects of early-stage NSCLC. Although there are still barriers yet to conquer, liquid biopsy is a hopeful detection means to be put into clinical use.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is a major global health concern, ranking as the second most commonly diagnosed cancer and the leading cause of cancer-related deaths in 2020 [1]. It can be classified into two main types: small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC). NSCLC accounts for 85% of all cases and is the predominant form of lung cancer [2]. Unfortunately, lung cancer is often asymptomatic in the early stages, with about 60% of patients presenting with distant metastasis at the time of diagnosis. This underscores the importance of an effective early screening methods for lung cancer. Even after surgical resection, early-stage NSCLC patients are still at risk of local or distant recurrence and metastasis [3]. To address this problem, a sophisticated recurrence risk evaluation system and a well-organized personal adjuvant therapy (ADT) strategy are necessary to optimize the outcomes for these patients.
Liquid biopsy is a genetic detection technology that takes the sampling of analytes from biological fluids, usually blood, but also other clinical secretions such as urine, ascites, and cerebrospinal fluid. Fragmented particles or dissociative cells originating from tumor cells’ growth or necrosis that carries genomic information are major subjects to the analysis, which helps us to extract information from the tumor and reveal key characteristics of it [4]. It is not only capable of clarifying the status of cancer and tumor load but also suitable to depict heterogeneity landscape and dynamic genetic patterns of cancer during treatment, which is hard to obtain from clinical tissue biopsy. Liquid biopsy is also applicable to practical utility, for its noninvasiveness, repeatability, real time surveillance and easy to access [5]. These advantages drew the attention of researchers to further explore its potential applications in clinical fields. Currently, different biomarkers in peripheral blood have been explore. Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), exosomes, microRNAs (miRNA), peripheral blood circulating RNA, tumor-educated blood platelets (TEPs), and circulating tumor vascular endothelial cells (CTECs) are typical detection subjects to liquid biopsy. CtDNA, CTCs and exosomes are the hottest biomarkers in the field of NSCLC since inception to May 2022 [4].
With the advancement of detection technology and refinement of clinical models, liquid biopsy has spread its utility in the whole diagnosis-treatment procedure of NSCLC. With the improvement of screening methods and improvement on people’s awareness of physical examination, the ratio of early stage lung cancer increased dramatically. Meanwhile, this phenomenon also brought about technical challenges to liquid biopsy in finding genetic mutations within less concentration, as early stage tumors shed little DNA fragments into blood stream. This review is going to introduce the mainstream detection methods in liquid biopsy and summarize the development and barriers of liquid biopsy in clinical applications on resectable early-stage NSCLC without distant metastasis staging from TNM I to IIIA.
2 Definition and detection methods for liquid biopsy biomarkers
2.1 Detection and biological characters of ctDNA
Circulating free DNA (cfDNA) was first found in human bloodstream in 1948 by Mandel and defined as degraded DNA fragments that are circulating freely in the bloodstream [6]. These shed fragments are usually generated from normal healthy leukocytes and stromal cells which have high proliferation rate. However, its relationship with tumor was revealed later by Shapiro in 1983 [7], as patients with cancer carried a general higher level of cfDNA in blood than healthy participants. These group of ctDNA that released from cancer cells were defined as circulating tumor DNA (ctDNA). Although ctDNA only contributes a little proportion to total cfDNA in early-stage NSCLC, it has manifested various talents in cancer diagnosis, prognostic evaluation and therapy management [8, 9].
The process of ctDNA’s release is deemed to be related with tumor cell’s death, such as apoptosis or necrosis, and sometimes ctDNA originates from active metabolism such as secretion when the cells were alive [10]. In practical analysis, it is hard to distinguish ctDNA from other cfDNA, because genetic aberration varies among cancer types and is even heterogeneous in each individual. Even though, ctDNA still share some typical and distinct biological features compared with the ones from healthy tissue. The length of cfDNA centers around 166 bp, similar to the length of DNA around a nucleosome plus its linker, as the result of histone’s protection from caspase-dependent endonuclease. Circulating DNA from tumor tissue is observed to have a different fragment pattern, approximately 23 bp shorter than others, but the reason remains unknown [11]. In circulation system, ctDNA faces a fast clearance via the kidneys, liver, and spleen. Based on the concentration analysis of multiple time point sampling, 35 min is determined to be the half-life of ctDNA in surgical resected lung cancer patients, which laid a solid foundation for dynamic surveillance [12]. With the advancement of detection techniques, quantification of ctDNA can even realize tumor volume prediction without invasive procedure. A linear relationship between plasma variant allele fraction (VAF) and tumor volume by CT volumetric analysis was constructed in a NSCLC cohort, among which a primary tumor burden of 10 cm3 is parallel to a mean plasma VAF of 0.1% [13].
Strong clinical value as it shows, there are plenty barriers in analyzing these circulating DNA. At each tumor cell’s death, only 0.014% of tumor’s DNA is shed into bloodstream [14]. An average concentration of cfDNA is estimated to range from 1 ~ 100 ng/ml in peripheral blood, while merely 0.1~1% of cfDNA comes from tumor tissue in early-stage patients, which is harsh for the sensitivity of detection technique, especially in early-stage patients with low tumor load. Besides, the background interference of circulating DNA from normal cells is another factor that influences the detection result. Somatic mutations that accumulate in hematopoietic cells with aging, termed clonal hematopoietic mutations of indeterminate potential (CHIP), may be misjudged as cancerous mutations [15]. To avoid false positive situations, CHIP-associated variant filtering or white blood cell control is necessary for hyper-sensitive detection approaches.
2.2 Detection methods for ctDNA
Unique characters of ctDNA as a tumor biomarker in peripheral blood can be classified into three major categories: (1) Genome mutations and structural variations; (2) Fragmentomic patterns; (3) Epigenetic characters. These biomarkers have found their own advantages in different areas of lung cancer management and their detection strategy also varies based on their disparate biological features (Table 1).
Before genomic detection, preanalytical parameters from blood drawing to the storage of cfDNA extracts are critical factors directly influencing the outcome. The largest contamination is from the preanalytical degradation of white blood cells (WBC) which releases an overwhelming amount of genomic DNA into the sample compared with the small quantity of ctDNA. To reduce WBC’s interference, plasma is a better sample than serum because plasma sample minimize the WBC-origin DNA from WBC lysis during the clotting process. And EDTA has a lower cfDNA increase rate than heparin and citrates as an anticoagulant. Before separation of the sample from blood cellular components, the amount of cfDNA has no significant increase in the first 4 ~ 6 h, no matter in room temperature or 4 °C storage. But the quality of cfDNA extracts for quantification analysis can only be guaranteed under − 20 °C for 3 months. During these procedures, agitation should be avoided and it is recommended that freeze–thaw process be minimized up to 3 cycles [16, 17].
2.2.1 Genome mutations and structural variations
Currently, the technical platforms that meet the requirements of ctDNA detection are mainly based on PCR or Next-generation Sequencing (NGS). PCR-based methods include real-time quantitative polymerase chain reaction (RT-PCR), droplet digital PCR (dd-PCR), beads-emulsion-amplification-and-magnetics (BEAMing) and amplification refractory mutation system (ARMS) [8]. RT-PCR is the most standardized PCR methods, but with a relatively low sensitivity. Detection accuracy is improved in dd-PCR by dividing the PCR reaction in plenty of droplets containing nucleic acid template. As for BEAMing methods, PCR is conducted by magnetic beads for the water-oil single-molecule amplification reaction, which had relative high sensitivity at the cost of a complex workflow. ARMS utilized the match at 3’ end of the primer to identify targeted mutations [5, 18]. These methods have modest detection accuracy and low economic requirements, but can only interrogate known mutations, and neither copy number variants nor fusion genes can be identified. Methods based on second-generation sequencing (NGS) include Cancer Personalized Profiling by deep sequencing (CAPP-Seq) and tagged amplicon deep sequencing (TAM-Seq). The detection range of NGS is not limited to several simple mutations, but expands its field to fusion genes, insertion/ deletion and copy number variations (CNVs). However, with advancement in detection efficiency, its shortcomings lie in longer analytical time cycle and higher economic cost, and certain bioinformatics knowledge storage of detection equipment is in need [8, 19]. In clinical application, early diagnosis of lung cancer favors NGS to discover unknown mutations, while PCR is useful in postoperative monitoring or drug-resistant management based on candidate genes information from tissue biopsy.
2.2.2 Fragmentomic patterns
It is widely accepted that ctDNA is more fragmented than cfDNA from clonal hematopoietic cells. The length of these shorter cfDNA oscillates in a 10 bp periodicity, which is associated with nucleosome or protein complex wrapping and protecting DNA from cleavage. Furthermore, nucleosome positioning is another important piece of information extracted from fragmentomic patterns, which showed relations with tumor origin and can be revealed through nuclear chromatin micrococcal nuclease (MNase) sequencing assays. Indeed, the loss of nucleosome positioning on both sides of transcription starting sites (TSS) is necessary to properly express genes. NGS-based methods are capable of analysis on fragment’s length and distribution, while as for nucleosome positioning, bioinformatics analysis is inevitable and not applied in routinely diagnosis [20].
2.2.3 Epigenetic characters
DNA methylation, histone post-translational modifications, histone variants and chromatin remodeling complexes are the main studied epigenetic mechanisms in liquid biopsy. DNA methylation refers to addition of a methyl group to cytosine in cytosine-phosphate-guanine (CpG) island, while histone post-translational modifications adds specific biochemical modifications on histone tails to regulate gene expression. Histone variants regulate chromatin remodeling and histone post-translational modifications with a few variant amino acids. In comparison, chromatin remodeling complexes remove, relocate or shift histones to regulate the nucleosome structure [21]. Among them, DNA methylation is the mostly studied biomarker in lung cancer management, which is also validated to provide information on early screening, prognosis and therapy response with robust performance.
Methylation on DNA is usually catalyzed by DNA methyltransferase enzymes (DNMTs) which transfer methyl group on S-adenosylmethionine (SAM) to the 5-position carbon of a cytosine ring. These methylations happen predominantly in CpG dinucleotides, 60–80% of which are generally methylated in healthy genome [22]. Methylation regulates specific gene expression in two biological mechanisms. Hypermethylation on gene promoter directly silences transcription, while methylation on methyl-CpG-binding proteins (MBP), which recruit DNMTs and histone deacetylases (HDAC), leads to chromatin conformation changes that further repress gene transcription [21]. DNA methylation enjoys several specific biological characters that capacitates itself as a favorable biomarker. DNA methylation is stable and homogenous in specific type of cancer with little individual differences and methylation aberrations appear at an early stage of cancer development [23, 24]. Such preponderance set the stage for DNA methylation to track cancer’s minute trails in bloodstream.
Technologies for ctDNA methylation assay can be roughly categorized into bisulfite conversion-based method, restriction enzyme-based method and enrichment-based method. Methylated cytosines are screened out through deamination process of bisulfite salts, while unmethylated cytosines without protection are transferred into uracil. Follow-up PCR on specific CpG sequencing or WGS towards global DNA methylation profiling are conducted based on different detection strategies, such as methylated CpG tandems amplification and sequencing (MCTA-seq) or whole-genome bisulfite sequencing (WGBS) [25, 26]. However, the toughest problem on this approach is degradation of DNA resulting from bisulfate conversion. To solve this problem, bisulfite conversion-free methods come into being. Restriction enzyme-based method is designed based on methylation’s protection effect from restriction endonuclease on specific genome sequencing. Methylation restriction enzymes (MREs) such as such as HpaII, Hin6I and AciI, are used parallelly in MRE-seq, but the outcome is strictly limited to the availability of enzyme recognition sites and the extent of MRE’s digestion. Antibodies specific for 5mC and methyl-binding protein MECP2 are important choices for enrichment-based method. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) [27] and modified methyl-CpG binding domain protein capture sequencing (MBD-seq) [28] are classical detection models in this field. After refinement, both of them can offer a resolution to methylation landscape under a minimum DNA input of 1–10 ng.
In comparison, bisulfite conversion-based WGBS is still the gold standard for comprehensive and unbiased whole-genome DNA methylation profiling, but bears heavy work load and economic burden. A relative higher level of DNA input is required due to the destructive effect of bisulfate. The availability of MRE-seq is limited to the recognition sites of restriction enzymes and possible incomplete digestion causes further false positive situations. CfMeDIP-seq and MBD-seq are economic friendly, but impotent to single-base-pair resolution or coverage of unmethylated CpG sites [29].
2.3 Detection and biological characters of CTCs
CTCs are defined as tumor cells that have been sloughed from the primary tumor and are swept away by the circulatory or lymphatic systems [30]. CTCs are able to carry out information at many levels, including presence of CTCs, CTC count, immuno-cytochemistry, genomic, transcriptomic, and proteomic analysis [31]. Different from primary tumor cells, CTCs manifest EMT transition properties and stemness features to adapt to the environment in circulation system from primary focal lesion. CTCs have a general short half-life time of 1.0-2.4 h. Under the attack of body immune system and survival pressure from anoikis and bloodstream shearing forces, or even an assault from targeted drugs, less than 0.01% of CTCs survive in the journey and lead to a new blood-borne metastasis as “seeds” of primary tumor. Same to ctDNA, CTCs are also low in abundance and patients with cancer seldom carries CTCs more than 10 cells per milliliter [32]. Although such characters hinder the development of CTCs analysis, CTCs offer an opportunity to information at the DNA, RNA and protein levels. Multiple dimensions of detection grants possibilities for its future development. In previous studies, CTCs have demonstrated great values in early screening and prognostic management among various cancer types, including breast cancer and lung cancer [33].
2.4 Detection methods for CTCs
Simple component separation of plasma is required before cfDNA analysis, but CTCs isolation is a tough assignment as the prerequisite and foundation for downstream CTCs analyses. Different CTC isolation techniques have been developed, which can be generally divided into physical methods and biological methods. Physical methods are based on individual physical characters between CTCs and blood cells, such as size, density, electric charge, migratory capacity, and deformability to filter CTCs. And biological methods mainly count on antigen-antibody binding to select CTCs by recognizable tumor-specific biomarkers [34] (Table 1).
Physical separation method of CTCs faces lots of difficulties in development, among which size and density are the most favorable features. Although CTCs own an average higher size than normal blood cells, an apparent overlap between interfering leukocytes and small size CTCs is not negligible. Besides, CTCs’ broad heterogeneity in physical size across cancer types is also observed in previous studies [35]. Size-based separation strategies are grossly categorized into two mainstream technologies: membrane microfilters and size-based microfluidic CTC sorting devices [36]. With continuous refinement and technological honing, CTCs capture efficiency ranges from 47 to 98% with blood samples no more than 7.5ml in different size-based researches [35]. Another physical methods use specific density of RBCs, leukocytes, and cancer cells to centrifugate target components, among which silicone flotation technique and isopycnic density gradient centrifugation are commonly used classical methods [37]. OncoQuick [38] and AccuCyte-CyteFinder system [39] are popular density-based CTCs detection platform applied in current clinical researches and their average recovery rate reached 87% and 90% respectively, although large cohort study and lung cancer specific research are yet to accomplish. Physical separation method produces label-free, unmodified viable cells that can be applied in downstream analysis, but this strategy faces inference from leukocytes or other cells that should not be ignored.
Another strategy to discriminate CTCs is biological property-based technology which identifies the biomarkers CTCs produces under the process of specific physiological activities with the help of antibody–antigen interaction. However, no universal CTC antigens have been identified yet [37]. CTCs from epithelial malignancies are likely to maintain original phenotype, as a result, epithelial markers such as the epithelial cell adhesion molecule (EpCAM) and cytokeratins are suitable for detection [40]. As the most commonly used biomarker, EpCAM antibody-coated ferromagnetic beads are applied in the only FDA-approved clinical device, CellSearch system, with removal of background CD45 + white blood cells. Even so, EpCAM is not a panacea for all cancer types. EpCAM-negative cell group even takes up a larger quantity than EpCAM-positive ones in NSCLC [41]. In the process of invasion, part of carcinoma cells experience epithelial-to-mesenchymal transition (EMT) to attain stronger migration ability, at the cost of epithelial phenotype. As a result, EpCAM-based enrichment will inevitably miss CTCs with higher EMT extent. Furthermore, negative-enrichment methods are taken into consideration due to its advantage of label-free viable CTCs. Negative immunomagnetic CD45 + white blood cell enrichment and red blood cell lysis are two common practices in research [37].
In clinical utility, combined approach is a favorable choice for CTCs analysis. CTC-iChip, an inertial focusing-enhanced microfluidic CTC capture platform, combined size-based filtration and immunomagnetic separation with positive selection and negative depletion modes and overcame previous CellSearch system in a pan-cancer cohort including lung cancer [42].
2.5 Detection and biological characters of exosomes
Exosomes are endosomal-origin vesicles ranging from 40 to 160 nm in diameter. Proteins, messenger RNAs (mRNAs), microRNAs (miRNAs), and lipids enriches in endosome, enclosed by a stable lipid bilayer, which mediates intracellular transfer of information in body fluids [43]. Exosome biogenesis begins with invagination of endosomal limiting membranes and ultimately forms multi-vesicular bodies (MVBs) after the process of early sorting endosomes and late sorting endosomes. Vesicles inside of MVBs, named intraluminal vesicles (ILVs), becomes exosomes in body fluid by the fusion of MVBs with plasma membrane [44]. Exosomes in body fluids have a general large abundance about 109 particles/mL and cancer cells secrete tenfold of exosomes than normal cells [45, 46]. As a result, exosomes with tumor information have easier access to obtain. Besides, exosomes reflect biological information from living cells, while information carried by ctDNA is mainly from apoptotic or dead tumor cells [46].
In lung cancer, tumor-derived exosomes play an important role in tumor development and metastasis through immune system evasion, epithelial-mesenchymal transition and angiogenesis. Exosomes from lung cancer impair immune system with immunosuppressive programmed death-ligand 1 (PD-L1) which inactivate T cells via extracellular domain and cause CD8 + T cell disfunction [47]. Furthermore, tumor-derived exosomes and microvesicles produce high level transforming growth factor (TGF)-β1 and miR-23a under hypoxia environment, which is a common situation in solid tumors. MiR-23a specifically targets the expression of CD107a in NK cells, inducing inhibition in cytotoxicity [48]. Second, exosomes from mesenchymal cells manifest upregulated β-catenin with suppressed expression of E-cadherin and vimentin as a reflection to this EMT status [49]. Moreover, exosomes from metastatic lung cancer cells with EMT signals successfully induced increased level of vimentin on normal bronchial epithelial cells, suggesting exosomes are key drivers to EMT and transform cancer cells into a more aggressive phenotype [50]. In another study, overexpression of miR-210 in exosomes from lung cancer was found to induce fibroblast reprogramming into cancer-associated fibroblasts through the regulation of JAK2/STAT3 signaling pathway and TET2 in recipient fibroblasts, the result of which enhanced angiogenesis due to cancer-associated fibroblasts’ acceleration to this process [51]. Without any doubt, exosomes spur the progression and metastasis of lung cancer, granting it the ability to become a first-class liquid biopsy biomarker.
2.6 Detection methods for exosomes
Exosome are heterogeneous in size and its cargo also varies between each of those. Exosome separation and enrichment are essential for statistical analysis and clinical translation. Common enrichment tools are listed as below. (1) Ultracentrifugation-based separation is rendered gold standard in exosome separation, including differential ultracentrifugation and gradient density ultracentrifugation. In comparison, gradient density ultracentrifugation provides better purity, but is more time-consuming and requires more samples to operate [52]. (2) Size-based separation shifts fixed-range diameter exosomes via specific pore sizes, but this method faces disadvantage of low yield. Combining multiple strategies or upgrading detection platform can effectively improve the efficiency. A size-based exosome total isolation chip (ExoTIC) by Liu enriches exosomes with multiple nanoporous membranes and laid the foundation for clinical testing from fingerprick quantities (10–100µL) of blood [53]. Another innovative design of perpendicular state of flow direction and filtration direction, named tangential flow filtration (TFF), solved the problem of pore clogging and developed a microfluidic tangential flow filtration device, Exodisc-B, which restrained the detection into 10–40 min with 30–600 µL volume blood [54]. (3) Immunoaffinity enrichment are also applied in exosome separation. Anti-CD81 functionalized microfluidic chip manufactured by Zhang afforded a detection limit of 50 µL [55]. Kang’s extracellular vesicles on demand (EVOD) chip applied dithiothreitol release of isolated EVs for downstream analysis and selected 76% more exosomes with EGFR in NSCLC patients than healthy donors, referring initial value in lung cancer early screening [56]. (4) Lipid-based separation is another methods to enrich exosomes, which targets on lipid molecules or the molecules absorbed on the exosome membrane. Wan’s team invented a nanoscale extracellular vesicle isolation platform by a surface-conjugated lipid nanoprobe with one end attached into membrane and the other providing solubility [57]. This kind of separation methods are characterized by its enrichment speed. The technique mentioned above only takes 15 min for isolation and another enrichment focusing on exosome phospholipids achieved a capture efficiency of 96.5% within 5 min [58]. (5) Acoustic-based isolation methods utilize ultrasonic waves to segregate exosomes based on their physical properties such as size and density. Wu integrated acoustics and microfluidics modules to reach a label-free, contact-free isolation. These modules are able to separate exosomes from an extracellular vesicle mixture with a purity of 98.4% [59] (Table 1).
3 Clinical applications of liquid biopsy
3.1 Early diagnosis
Early detection is necessary for potential lung cancer patients to reduce mortality, as the 5-year overall survival rate is closely related to the TNM stage when first diagnosed [1] .Present recommended detection strategy is low dose CT (LDCT) for smokers and former smokers who age above 55 with a 30 pack-year smoking history. Many randomized clinical trials (RCTs) supported the status of LDCT in those high-risk populations and numerous lung cancer screening guidelines, such as the National Comprehensive Cancer Network (NCCN) [60] and American Society of Clinical Oncology (ASCO) [61], recommend lung cancer screening by LDCT for individuals with high risk. Among those RCTs, the National Lung Screening Trial (NLST) and Nederlands-Leuvens Longkanker Screenings Onderzoek (NELSON) were the largest and proved clear benefits for a specific population. NLST recruited 53,454 high-risk participants and a decrease of 20% in lung cancer-related mortality was found in the LDCT group during a 3-year follow-up procedure [62]. NELSON trial was conducted among 15,789 smokers and the interval of screenings and follow-up time were elongated, the result of which exhibited similar outcomes favoring LDCT conducted once every several years to achieve improvement in survival rate [63]. However, LDCT is far from perfect for lung cancer screening. (1) Extremely high false positive rate is the most vital imperfection for LDCT. In the NLST, the false positive rate ranged from 15.9 to 27.2% in each year’s test [62]. The NELSON noted a lower false positive rate, but still, fluctuate at a relatively high level [63]. (2) Overdiagnosis and unnecessary invasive procedure are the unwanted consequences close behind. A meta-analysis summarized overdiagnosis in LDCT screening, an estimation of the overdiagnosis rate reached 67.2% at most [64]. In NLST, 1.7% of those screened participants faced invasive procedures, 0.4% of false positive participants underwent at least one complication, and 0.1% of them even experienced death after an invasive diagnostic procedure [62]. (3) Even though LDCT has already decreased the radiation dose to 0.65 mSv to 2.36 mSv per screening, a regular reexamination can bring substantial injury to patients [64]. The lifetime risk of major cancers related to the radiation from 10 annual LDCT scans was 0.26 to 0.81 in every 1000 people screened [65]. (4) Psychosocial harms from the screening may also reduce quality of life. Individuals receiving true-positive results or indeterminate results experienced most anxiety and distress [66]. But this outcome requires better large cohort RCTs to validate its reliability.
Liquid biopsy’s sensitivity and convenience supported its utility in lung cancer early detection, while its invasiveness and applicability partly solved the above difficulties in LDCT. However, biomarkers carrying tumor information are quite low in density in peripheral blood, as a median cfDNA concentration is as low as 8.64 ng/ml in stage I NSCLC [67]. As a result, detection tools need to be sensitive enough to finish this job. Tracking Non-Small-Cell Lung Cancer Evolution Through Therapy (Rx) (TRACERx) study utilized SNVs in ctDNA as a biomarker to refer a cancerous status under NGS among 96 early-stage NSCLCs. As lung squamous cell carcinomas (LUSCs) were more necrotic than lung adenocarcinomas (LUADs), the detection efficiency of LUSCs are significantly higher than LUADs [13]. With the advancement in technology, researchers consistently pushed the detection threshold to allow more possibility to discern patients with early stage cancers. After the pan-cancer screening platform, Cancer-SEEK, which targeted on eight circulating protein biomarkers [68] and tumor-specific mutations and a targeted error correction sequencing, named TEC-Seq [69], cancer personalized profiling by deep sequencing (CAPP-Seq) was honored as the most accurate ctDNA mutation detection platform with the highest sensitivity for early stage NSCLC. CAPP-Seq was an NGS-based method which combined optimized library preparation methods with a multiphase bioinformatics approach. It provided 100% sensitivity with stage II-IV tumors, but stage I lung cancer only had 50% sensitivity at a specificity of 96% [70]. Although marvelous performance it had manifested, only 13 patients were involved in this article for verification. Chabon’s team refined the recovery and enrichment procedure of CAPP-Seq to develop a new machine-learning method, named lung cancer likelihood in plasma (Lung-CLiP), which distinguished cancers at a sensitivity of 67.3% with the specificity of 80% in a cohort containing 104 patients with NSCLC and 56 risk-matched controls [71]. Lung-CLip also achieved 63% of sensitivity under 80% specificity in stage I subgroup. Apart from finding patients carrying lung cancer, ctDNA mutation can further predict tumor volume based on mutant allele frequency. TRACERx first found a linear relationship between log-transformed clonal plasma VAF and log-transformed tumor volume under CT [13]. Abbosh systematically reviewed this discovery as mean clonal mutant allele frequency of 0.1% corresponded to 10cm3 tumor burden and roughly forecasted a T1c stage [72]. Another study integrated data from several researches and concluded 0.21 haploid genome equivalents per plasma ml referred to 1 cm3 of tumor volume. 0.014% of a tumor cell’s DNA shedding rate at each cell’s death was also calculated as a microscopic evidence for its ability to reflect tumor burden [14].
Methylation is another sensitive biomarker for early screening, for its appearance is anterior than gene alteration, even in a precancerous stage, which offered more chances to distinguish early stage patients. Recent research compared detection performance in pan-cancer early detection between different cfDNA features. Considering background removal with white blood cell pairing, best detection strategies were applied in each features analysis and whole genome methylation overwhelmed others in clinical limit of detection, suggesting methylation kept the lowest detection threshold in early detection among various cfDNA approaches [73]. A refined bisulfite-conversion based methylation detection approach named methylation on beads quantitative methylation-specific PCR (MOB-qMSP) was applied in Chinese NSCLC cohort, the combination of CDO1, TAC1, and SOX17 was evaluated as the best indicator for early detection with sensitivity and specificity at 90% and 71% in a cohort of 163 stage I NSCLC patients and 83 benign controls [74]. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) is another methylation approach, which utilized bisulfite-free technique with whole-genome methylation. With these technical support, cfMeDIP-seq achieved better detection performance under random forest model with high sensitivity and specificity of 91.0% and 93.3% within a cohort of 67 cancers with 59.7% of stage I and 30 normal controls, noting methylation’s advantages on early detection and validating its potential in NSCLC [75].
CTCs’ utility in NSCLC early screening has also been examined, but the outcome showed less competence than other popular biomarkers. A filtration-based detection technique, isolation by size of epithelial tumor cells (ISET), was assessed in malignancy discernment in lung cancer. When 25 CTC count was delineated as the threshold, sensitivity in malignant group reached 89% without false positive individuals among 60 neoplastic patients and 17 benign controls [76]. However, the following multi-centered research in France gave out pessimistic results. A 3-year ISET screening was conducted in a multi-centered high-risk population attempting to check its talent to be a regular screening examination, among which 614 participants took part in and only 19 turned out to be lung cancer patients verified by pathology diagnosis. As the outcome, the sensitivity of CTC detection for lung cancer detection was just 26.3%, far from the standard of a clinical screening method [77]. The deviation of the two researches originated from sufficient preliminary experiments. The formal research only took 60 malignant samples to validate CTC’s ability, but the following screening trial referred that CTC was not up to standard for large population screening and CTC required more evidence and technique support to enhance its ability. However, it is too early to deny CTC’s value in early detection, since its convenience in analysis and economic benefits are friendly to clinical utility and the combination of CTCs with other biomarkers is worth the wait.
MiRNA in exosomes has performed favorable availability in NSCLC early detection. Jin’s research identified specific miRNA profiles of adenocarcinoma and squamous cell carcinoma (SCC) in early NSCLC diagnosis from peripheral blood. Combination miRNA panels for NSCLC scored a sensitivity of 80.25% and a specificity of 92.31% in the stage I NSCLC cohort with 47 malignant samples and 13 healthy ones. The research further developed miR-181-5p, miR-30a-3p, miR-30e-3p, and miR-361-5p adenocarcinoma-specific panel, and miR-10b-5p, miR-15b-5p, and miR-320b SCC-specific panel respectively, both of which exhibited high AUC value over 0.91 [78].
These biomarkers all exhibited great performance in early screening of NSCLC. The current trend is to find out a clinically friendly combination which is not only sensitive but also owns real benefits than traditional strategy.
Under many early detection strategies, we intended to compare the practicability of liquid biopsy strategy with conventional detection methods, including LDCT and serum tumor markers (Table 2). Medical imaging examination is the most commonly used way to find a potential pulmonary cancer in clinical practice. NLST [62] and NELSON [63] trials has proved LDCT can decrease cancer-related mortality by 20% but with a 96.4% false positive rate. Another meta-analysis summarized the efficiency of LDCT on lung cancer detection from English-language articles published through May 2019. 13 studies including more than 75,000 participants were involved and LDCT’s sensitivity ranged from 59 to 100%, while specificity from 26.4–99.7% [64]. LDCT’s accuracy ranged dramatically due to heterogeneity of eligibility criteria, screening protocols or follow-up length. LDCT often requires years of regular follow-up to obtain a good diagnostic ability, and the missed diagnosis and misdiagnosis rate of a single examination are very high [79]. A large-scale RCTs including 6538 patients underwent 4 rounds of LDCT screening in 5.5 years, taking an additional 2 year of follow-up from the national cancer registry as gold standard, whose result showed 59.0% of sensitivity and 95.8% specificity [80]. In comparison, liquid biopsy had higher detection efficiency than traditional LDCT, however no liquid biopsy research was applied on a large-scale screening cohort similar with the ones on LDCT. As an early screening means, the detection cycle remained suspensive for liquid biopsy, yet a single round of detection had proved favorable outcome in previous articles [71, 75, 76, 78] which indicated a good potential of a powerful clinical tool for lung cancer early detection. Apart from that, serum tumor markers are also frequently-used in clinical practice. Classical serum biomarkers, such as neuron-specific enolase (NSE) and carcinoembryonic antigen (CEA), are pan-cancer specific and tend to be hyposensitive in NSCLC [81]. A prospective research calculated the diagnostic value of CEA, cancer antigen (CA125), squamous cell carcinoma (SCC), cytokeratin 19 fragment antigen21-1 (CYFRA 21 − 1) and NSE in NSCLC. Single marker had low sensitivity, ranging from 22 to 76%. CYFRA 21 − 1 and CA125 appeared to be the most sensitive combination in the cohort which involved 142 localized NSCLC and reached sensitivity of 83.8% [82]. In conclusion, although traditional detection means had passable accuracy in NSCLC early detection, non-inferior outcomes could be achieved in earlier populations by liquid biopsy-based methods.
3.2 Prognosis Judgment
Liquid biopsy is useful for early-stage lung cancer screening and has great value in prognostic performance. Minimal residue disease exists after the initial therapy and represents the possibility of primary cancer recurrence or advancement. Detection of these MRD required liquid biopsy to reveal timely intervention, including detection of ctDNA, CTCs, and exosomes. Newman explored CAPP-Seq, a sensitive method covering multiple groups of somatic alterations, which designed a selector with biotinylated DNA oligonucleotides targeting regions of cancer interest. This technology set a milestone in MRD monitoring, as it challenged the stage of radiography in recurrence prediction with better prediction accuracy in specific individuals and made up for the imaging deficiencies to help distinguish residual tumor or postradiotherapy inflammation which had similar radiographic features [70]. In another cohort containing 40 early-stage lung cancer patients conducted by Chaudhuri, evaluation method by CAPP-Seq prevailed over radiographic progression by RECIST 1.1 criteria in 72% of patients, earlier by a median of 5.2 months. Additionally, Chaudhuri set the MRD landmark at the first posttreatment blood draw within 4 months of treatment completion. This landmark had accuracy up to 96.87% for prediction of progression among 32 NSCLC patients in the following 72 months after the landmark [83]. TRACERx study defined the SNV threshold for the first time in MRD monitoring. Taking 2 SNVs as the standard, 13 out of 14 patients experiencing relapse within the first two years after surgery were rendered positive before or overlapped with the relapse timepoint. While only 1 out of 10 progression-free patients was classified wrong [13]. So far, the application of postoperative ctDNA to predict survival has proved its feasibility, but large-scale verification is still lacking. Many researchers afterward threw themselves into the validation work of ctDNA in MRD, looking forward to exploring different panels and expanding this application on patients in earlier stages. Among them, the LUNGCA project contained 67% of stage I lung cancer patients with a total of 330 NSCLC participants. Researchers also shifted the MRD landmark to 1 month after the surgery and got a sensitivity of 80.8% and specificity of 83.8% using MinerVa MRD assay. In this cohort, ctDNA-based MRD owned higher relative contribution to survival prediction than TNM stage [84].
In the field of CTCs, EpCAM-based CellSearch system and filtration-based ISET technology are major methods in research on the prognostic value. In researches based on CellSearch system, blood sample in peripheral vein before surgery and the one from pulmonary vein taken during the operation were usually applied in survival analysis. Among 30 lung cancer patients receiving thoracotomy, there were 16.7% and 50.0% of positive rate respectively in those two kinds of samples, but no significance was found in recurrence or overall survival with a median follow-up of 13 months [85]. Apart from this one, another CellSearch project adopting more relaxed positive criteria with similar design showed positive rate around 20% each but revealed significance in DFS and 3-year OS with CTCs in both samples from 30 NSCLC patients staging from I to IIIA [86]. On the other hand, the CTC counts of more than 50 underwent ISET method from the preoperative blood sample was significantly associated with shorter OS and DFS for stage I and II patients, proved in a 28 NSCLC patients’ cohort [87]. Hofman V also discovered similar conclusion in another 174 stage I to stage II NSCLC patient cohort, that the presence of CTCs in preoperative peripheral blood detected by ISET was a significantly independent prognostic factor for shorter DFS [88]. These result revealed CTCs’ potential in prognostic performance, but the outcome varied in different methods. This discrepancy may reflect limitations in EpCAM-based method, as EpCAM not always kept a high level on CTCs, which was verified by the differentiated rate of identification to diverse stages of cancerous participants [88]. The prognostic efficiency of MRD monitoring in CTCs required further validation or better detection strategy in early-stage NSCLC.
Exosomes are also useful biomarkers in predicting patients’ prognostic performance. Dejima’s team revealed an upregulation of the exosomal miR‑21 and miR‑4257 levels in NSCLC patients with recurrence through microarray‑based expression profiling. Afterward, worse disease‑free survival (DFS) rates up to 30 months were found to have a close correlation with high levels of either miR‑21 or miR‑4257 in another independent cohort with 195 NSCLC patients [89]. Similarly, Liu utilized qPCR array panel selected 9 candidate miRNAs and generated an optimized combination of exosomal miR-23b-3p, miR-10b-5p, and miR-21-5p as prognostic indicators for NSCLC patients. In a 196 NSCLC cohort, this miRNA combination improved the survival predictive accuracy with the AUC of the clinical variables model from 0.88 to 0.91 at the time of 12 months [90]. Additionally, upregulation of Let-7a-5p which induced cancer cell death through BCL2L1-mediated PI3Kγ signaling pathway, also showed potential prognostic value in lung cancer patients, but this Let-7a-5p biomarker still lack a direct validation to survival of lung cancer patients [91, 92].
At present, many pieces of research have proven the prognostic value of liquid biopsy including ctDNA, CTCs, and exosomes in lung cancer. This kind of molecular recurrence also has a lead-time effect than traditional radiological monitoring. In Chaudhuri’s CAPP-Seq analysis, 72% of patients got a median of 5.2 months ahead of radiographic progression [83]. And the median interval in TRACERx cohort was 70 days, ranging from 10 to 346 days [13]. A meta-analysis on ctDNA detection integrated 9 eligible articles and calculated an average lead time of 179 days than radiology progression [67].
Although scientists have found numerous biomarkers suitable for prognostic assessment, there are many problems yet to solve before its popularization in the clinic. First, few regulations are posted to define a unified time point or optimal detection technology for follow-up visits. Apart from exploring the best MRD monitoring strategy, former cohorts also required long-term prognostic outcomes. The mismatch of ctDNA-positive with current progression may result from a short follow-up period and the relationship of MRD with long-term DFS or OS in early-stage resectable NSCLC is granted with many expectations [93].
3.3 Predictive value in ADT
Although adjuvant therapy was proven to bring substantial benefits to early-stage patients, its benefits and concomitant harm to patients are worth balancing. A meta-analysis by the lung adjuvant cisplatin collaborative group indicated a 5-year absolute benefit of 5.4% from chemotherapy in 3485 completely resected NSCLC patients’ group. However, the benefits varied considerably with the clinical stage and there are even negative benefits for very early-stage patients [94]. Additionally, highly toxic adjuvant chemotherapies also increase the risk of non-cancer death, and its benefits ought to balance with other risks, like permanent hearing loss or kidney damage, nausea, and fatigue [95]. Hence, new criteria for selecting patients with little need for adjuvant therapy are in need to reduce these unnecessary side effects.
Current studies were limited to ctDNA, similar research on CTCs or exosomes rarely focused on resectable early-stage patients. In early-stage NSCLC adjuvant therapy prediction value assessment, the biomarker mainly concentrated on ctDNA. Few pieces of research explored this kind of therapeutic efficiency through CTCs and exosomes. In the field of ctDNA, Chen first reported differences in RFS regarding to patients receiving adjuvant therapy or not in MRD positive group in resectable NCSLC and the median RFS was 268 days in receiving ADT versus 111 days in not receiving ADT [12]. This result refers to the possibility of ctDNA indicators to decide ADT management, instead of just consulting TNM staging. Subsequent large cohort studies verified this hypothesis with consistency that postoperative ctDNA-positive patients would have longer RFS if receiving adjuvant therapy [84, 96,97,98]. But the adjuvant therapy strategy in MRD negative group varied, as some revealed that RFS was not correlated with receiving ADT or not, while others found ADT had real survival benefits in those patients. The barrier on predictive value is also apparent, the MRD-negative cohort lacked a definite conclusion on ADT’s benefits. As for these patients, ADT might not result in shorter survival, but safety and life quality were also important to patients if they had no apparent survival benefits.
4 Conclusions
Liquid biopsy is a versatile tool in lung cancer management. With the advancement of detection technology, diverse genomic detection tools emerged and liquid biopsy has manifested promising talents in various clinical scenarios. Its utility in early screening improved the accuracy to identify latent lung cancer, in prognostic performance augmented the ability to predict recurrence risk more than TNM stage, in predictive value acted as a predictive factor to judge adjuvant therapy’s benefits. However, there are still many problems waiting for us to conquer, before we propel its application into practical clinical use.
Availability of data and materials
Not applicable.
References
Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet. 2021;398:535–54.
Demicheli R, Fornili M, Ambrogi F, et al. Recurrence dynamics for non-small-cell Lung cancer: effect of surgery on the development of metastases. J Thorac Oncol. 2012;7:723–30.
Li W, Liu JB, Hou LK, et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer. 2022;21:25.
Liu C, Xiang X, Han S, et al. Blood-based liquid biopsy: insights into early detection and clinical management of lung cancer. Cancer Lett. 2022;524:91–102.
Mandel P, Metais P. [Nuclear acids in human blood plasma]. C R Seances Soc Biol Fil. 1948;142:241–3.
Shapiro B, Chakrabarty M, Cohn EM, et al. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116–20.
Zhao H, Chen KZ, Hui BG, et al. Role of circulating tumor DNA in the management of early-stage lung cancer. Thorac Cancer. 2018;9:509–15.
Filipska M, Rosell R. Mutated circulating tumor DNA as a liquid biopsy in Lung cancer detection and treatment. Mol Oncol. 2021;15:1667–82.
Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–76.
Mouliere F, Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A. 2015;112:3178–9.
Chen K, Zhao H, Shi Y, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res. 2019;25:7058–67.
Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage Lung cancer evolution. Nature. 2017;545:446–51.
Avanzini S, Kurtz DM, Chabon JJ, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv. 2020;6:6.
Kilgour E, Rothwell DG, Brady G, et al. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37:485–95.
El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: preanalytical considerations. Clin Chim Acta. 2013;424:222–30.
Nikolaev S, Lemmens L, Koessler T, et al. Circulating tumoral DNA: preanalytical validation and quality control in a diagnostic laboratory. Anal Biochem. 2018;542:34–9.
Bhanothu V, Venkatesan V. Conventional polymerase chain reaction and amplification refractory mutation system-multi-gene/ multi-primer PCR in the diagnosis of female genital tuberculosis. Arch Microbiol. 2019;201:267–81.
Guibert N, Pradines A, Favre G, et al. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur Respir Rev. 2020;29:190052.
Keller L, Belloum Y, Wikman H, et al. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124:345–58.
Constâncio V, Nunes SP, Henrique R, et al. DNA methylation-based testing in Liquid biopsies as detection and prognostic biomarkers for the four major cancer types. Cells. 2020;9:624.
Hammerling MJ, Fritz BR, Yoesep DJ, et al. In vitro ribosome synthesis and evolution through ribosome display. Nat Commun. 2020;11:1108.
Widschwendter M, Jones A, Evans I, et al. Epigenome-based cancer risk prediction: rationale, opportunities and challenges. Nat Rev Clin Oncol. 2018;15:292–309.
Lianidou E. Detection and relevance of epigenetic markers on ctDNA: recent advances and future outlook. Mol Oncol. 2021;15:1683–700.
Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–7.
Wen L, Li J, Guo H, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res. 2015;25:1376.
Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–83.
Aberg KA, Chan RF, Shabalin AA, et al. A MBD-seq protocol for large-scale methylome-wide studies with (very) low amounts of DNA. Epigenetics. 2017;12:743–50.
Li S, Tollefsbol TO. DNA methylation methods: global DNA methylation and methylomic analyses. Methods. 2021;187:28–43.
Lin D, Shen L, Luo M, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. 2021;6:404.
Maly V, Maly O, Kolostova K, et al. Circulating tumor cells in diagnosis and treatment of lung cancer. In Vivo. 2019;33:1027–37.
Chen L, Bode AM, Dong Z. Circulating tumor cells: moving Biological insights into detection. Theranostics. 2017;7:2606–19.
Vasseur A, Kiavue N, Bidard FC, et al. Clinical utility of circulating tumor cells: an update. Mol Oncol. 2021;15:1647–66.
Ye Q, Ling S, Zheng S, et al. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18:114.
Hao SJ, Wan Y, Xia YQ, et al. Size-based separation methods of circulating tumor cells. Adv Drug Deliv Rev. 2018;125:3–20.
Ferreira MM. Circulating tumor cell technologies. Mol Oncol. 2016;10:374–94.
Bankó P, Lee SY, Nagygyörgy V, et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12:48.
Rosenberg R, Gertler R, Friederichs J, et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–8.
Campton DE, Ramirez AB, Nordberg JJ, et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer. 2015;15:360.
Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med. 2015;7:1–11.
Wang J, Lu W, Tang C, et al. Label-free isolation and mRNA detection of circulating tumor cells from patients with metastatic lung cancer for disease diagnosis and monitoring therapeutic efficacy. Anal Chem. 2015;87:11893–900.
Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147.
Li MY, Liu LZ, Dong M. Progress on pivotal role and application of exosome in Lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol Cancer. 2021;20:22.
Yu D, Li Y, Wang M, et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56.
Alipoor SD, Mortaz E, Varahram M, et al. The potential biomarkers and immunological effects of tumor-derived exosomes in lung cancer. Front Immunol. 2018;9: 819.
Cai X, Janku F, Zhan Q, et al. Accessing genetic information with liquid biopsies. Trends Genet. 2015;31:564–75.
Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–6.
Berchem G, Noman MZ, Bosseler M, et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology. 2016;5: e1062968.
Kim J, Kim TY, Lee MS, et al. Exosome cargo reflects TGF-β1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem Biophys Res Commun. 2016;478:643–8.
Rahman MA, Barger JF, Lovat F, et al. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016;7:54852–66.
Fan J, Xu G, Chang Z, et al. miR-210 transferred by Lung cancer cell-derived exosomes may act as proangiogenic factor in cancer-associated fibroblasts by modulating JAK2/STAT3 pathway. Clin Sci (Lond). 2020;134:807–25.
Royo F, Théry C, Falcón-Pérez JM, et al. Methods for separation and characterization of extracellular vesicles: results of a worldwide survey performed by the ISEV Rigor and Standardization Subcommittee. Cells. 2020;9:1955.
Liu F, Vermesh O, Mani V, et al. The exosome total isolation chip. ACS Nano. 2017;11:10712–23.
Sunkara V, Kim CJ, Park J, Automated F, et al. Label-free isolation of extracellular vesicles from whole blood for cancer diagnosis and monitoring. Theranostics. 2019;9:1851–63.
Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016;16:3033–42.
Kang YT, Hadlock T, Jolly S, et al. Extracellular vesicles on demand (EVOD) chip for screening and quantification of cancer-associated extracellular vesicles. Biosens Bioelectron. 2020;168:112535.
Wan Y, Maurer M, He HZ, et al. Enrichment of extracellular vesicles with lipid nanoprobe functionalized nanostructured silica. Lab Chip. 2019;19:2346–55.
Pang Y, Shi J, Yang X, et al. Personalized detection of circling exosomal PD-L1 based on Fe(3)O(4)@TiO(2) isolation and SERS immunoassay. Biosens Bioelectron. 2020;148:111800.
Wu M, Ouyang Y, Wang Z, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci U S A. 2017;114:10584–9.
Wood DE, Eapen GA, Ettinger DS, et al. Lung cancer screening. J Natl Compr Canc Netw. 2012;10:240–65.
Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for Lung cancer: a systematic review. JAMA. 2012;307:2418–29.
Aberle DR, Adams AM, Berg CD, et al. Reduced Lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409.
de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N Engl J Med. 2020;382:503–13.
Jonas DE, Reuland DS, Reddy SM, et al. Screening for lung cancer with low-dose computed tomography: updated evidence report and systematic review for the US preventive services task force. JAMA. 2021;325:971–87.
Rampinelli C, De Marco P, Origgi D, et al. Exposure to low dose computed tomography for Lung cancer screening and risk of cancer: secondary analysis of trial data and risk-benefit analysis. BMJ. 2017;356:j347.
Dunn CE, Edwards A, Carter B, et al. The role of screening expectations in modifying short-term psychological responses to low-dose computed tomography Lung cancer screening among high-risk individuals. Patient Educ Couns. 2017;100:1572–9.
Shen H, Jin Y, Zhao H, et al. Potential clinical utility of liquid biopsy in early-stage non-small cell lung cancer. BMC Med. 2022;20:480.
Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–30.
Phallen J, Sausen M, Adleff V. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9:9.
Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54.
Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early Lung cancer detection. Nature. 2020;580:245–51.
Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. 2018;15:577–86.
Jamshidi A, Liu MC, Klein EA, et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell. 2022;40:1537–1549.e1512.
Chen C, Huang X, Yin W, et al. Ultrasensitive DNA hypermethylation detection using plasma for early detection of NSCLC: a study in Chinese patients with very small nodules. Clin Epigenetics. 2020;12:39.
Qi J, Hong B, Tao R, et al. Prediction model for malignant pulmonary nodules based on cfMeDIP-seq and machine learning. Cancer Sci. 2021;112:3918–23.
Fiorelli A, Accardo M, Carelli E, et al. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg. 2015;99:1899–905.
Marquette CH, Boutros J, Benzaquen J, et al. Circulating tumour cells as a potential biomarker for lung cancer screening: a prospective cohort study. Lancet Respir Med. 2020;8:709–16.
Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23:5311–9.
Field JK, Duffy SW, Baldwin DR, et al. UK lung cancer RCT pilot screening trial: baseline findings from the screening arm provide evidence for the potential implementation of lung cancer screening. Thorax. 2016;71:161–70.
Horeweg N, Scholten ET, de Jong PA, et al. Detection of lung cancer through low-dose CT screening (NELSON): a prespecified analysis of screening test performance and interval cancers. Lancet Oncol. 2014;15:1342–50.
Chang L, Li J, Zhang R. Liquid biopsy for early diagnosis of non-small cell lung carcinoma: recent research and detection technologies. Biochim Biophys Acta Rev Cancer. 2022;1877: 188729.
Molina R, Filella X, Augé JM, et al. Tumor markers (CEA, CA 125, CYFRA 21 – 1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol. 2003;24:209–18.
Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–403.
Xia L, Mei J, Kang R, et al. Perioperative ctDNA-based molecular residual disease detection for non-small cell lung cancer: a prospective multicenter cohort study (LUNGCA-1). Clin Cancer Res. 2022;28:3308–17.
Okumura Y, Tanaka F, Yoneda K, et al. Circulating tumor cells in pulmonary venous blood of primary Lung cancer patients. Ann Thorac Surg. 2009;87:1669–75.
Crosbie PA, Shah R, Krysiak P, et al. Circulating tumor cells detected in the tumor-draining pulmonary vein are associated with disease recurrence after surgical resection of NSCLC. J Thorac Oncol. 2016;11:1793–7.
Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with Lung cancer is a new prognostic biomarker. Clin Cancer Res. 2011;17:827–35.
Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int J Cancer. 2011;129:1651–60.
Dejima H, Iinuma H, Kanaoka R, et al. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell Lung cancer. Oncol Lett. 2017;13:1256–63.
Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell Lung cancer. Oncotarget. 2017;8:13048–58.
Duan S, Yu S, Yuan T, et al. Exogenous Let-7a-5p induces A549 lung cancer cell death through BCL2L1-mediated PI3Kγ signaling pathway. Front Oncol. 2019;9: 808.
Zhang L, Hao C, Zhai R, et al. Downregulation of exosomal let-7a-5p in dust exposed- workers contributes to lung cancer development. Respir Res. 2018;19:235.
Li FQ, Cui JW. Circulating tumor DNA-minimal residual disease: an up-and-coming nova in resectable non-small-cell lung cancer. Crit Rev Oncol Hematol. 2022;179: 103800.
Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–9.
Chaft JE, Rimner A, Weder W, et al. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat Rev Clin Oncol. 2021;18:547–57.
Qiu B, Guo W, Zhang F, et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat Commun. 2021;12:6770.
Zhang JT, Liu SY, Gao W, et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non-small cell lung cancer. Cancer Discov. 2022;12:1690–701.
Moding EJ, Liu Y, Nabet BY, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small cell lung cancer. Nat Cancer. 2020;1:176–83.
Acknowledgements
Not applicable.
Funding
This work was supported by CAMS Innovation Fund for Medical Sciences(CIFMS)2022-I2M-C&T-B-120, Research Unit of Intelligence Diagnosis and Treatment in Early Non-small Cell Lung Cancer, Chinese Academy of Medical Sciences (2021RU002), National Natural Science Foundation of China (No.92059203, No.82072566), Clinical Medicine Plus X - Young Scholars Project, Peking University, the Fundamental Research Funds for the CentralUniversities(PKU2023LCXQ008), Beijing Natural Science Foundation (L222021) and Peking University People’s Hospital Research and Development Funds (RZ2022-03).
Author information
Authors and Affiliations
Contributions
Conceptualization: Yichen Jin, Kezhong Chen. Writing – original draft: Yichen Jin, Kezhong Chen. Writing – review & editing: Fan Yang, Kezhong Chen.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jin, Y., Yang, F. & Chen, K. An overview of current development and barriers on liquid biopsy in patients with early-stage non-small-cell Lung cancer. Holist Integ Oncol 2, 43 (2023). https://doi.org/10.1007/s44178-023-00066-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44178-023-00066-5