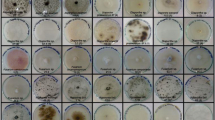
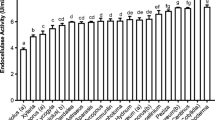
Abstract
This study aimed at the exploitation of lignocellulosic wastes for the evaluation of the newly isolated white-rot fungal strains enzymatic potential for bioethanol production. The isolates belonging to Basidiomycetes, Fomes fomentarius TMF2, Schizophyllum commune TMF3, and Bjerkandera adusta TMF1, could synthesize extracellular laccase and various hydrolase while growing on lignocellulosic waste materials. More specifically, for the first time, F. fomentarius TMF2 synthesized laccase using sunflower meal as a substrate. This substrate could stimulate B. adusta TMF1 for carboxymethyl cellulase and Avicelase production. The isolate B. adusta TMF1 was able to produce amylase during its growth on brewerʼs spent grain, which is up to now the best result reported for this activity of any B. adusta strain. Soybean meal was the most potent substrate for stimulating pectinase production by B. adusta TMF1 and S. commune TMF3. While growing on brewerʼs spent grain, B. adusta TMF1 and S. commune TMF3 produced high levels of xylanase. Spent coffee residues were for the first time tested as a substrate for hydrolase production by selected fungal species. Also, this is the first attempt where the produced enzymes by isolate B. adusta TMF1 were used for lignocellulose hydrolysis of brewerʼs spent grain and corn stover for bioethanol production, where under non-optimized conditions 0.94 g/L and 0.86 g/L of bioethanol could be produced, respectively. This study showed that novel white-rot fungal isolates, especially B. adusta TMF1, could grow on unexploited, low-cost lignocellulosic substrates and to produce biotechnological value-added products within environmental and economical accepted processes.
Graphical abstract


Similar content being viewed by others
Data Availability
Not applicable.
Code availability
Not applicable.
Abbreviations
- ANOVA:
-
Analysis of variance
- BSG:
-
Brewerʼs spent grain
- CMC:
-
Carboxymethyl cellulose
- CTAB:
-
Cetyl trimethyl ammonium bromide
- DNA:
-
Deoxyribonucleic acid
- DNS method:
-
Dinitrosalicylic acid method
- ITS:
-
Internal transcribed spacer
- MEA:
-
Malt extract agar
- NCBI:
-
The National Center for Biotechnology Information
- PCR:
-
Polymerase chain reaction
- rRNA:
-
Ribosomal ribonucleic acid
- SBM:
-
Soybean meal
- SCR:
-
Spent coffee residues
- SFM:
-
Sunflower meal
- SSF:
-
Solid-state fermentation
- WRF:
-
White-rot fungi
References
Serbent MP, Guimarães DKS, Drechsler-Santos ER, Helm CV, Giongo A, Tavares LBB (2020) Growth, enzymatic production and morphology of the white-rot fungi Lentinuscrinitus (L.) Fr. upon 2,4-D herbicide exposition. Int J Environ Sci Technol 17(5):2995–3012. https://doi.org/10.1007/s13762-020-02693-1
Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR (2014) Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol Rev 78(4):614–649. https://doi.org/10.1128/MMBR.00035-14
Elisashvili V, Kachlishvili E, Tsiklauri N, Metreveli E, Khardziani T, Agathos SN (2009) Lignocellulose-degrading enzyme production by white-rot Basidiomycetes isolated from the forests of Georgia. World J Microbiol Biotechnol 25(2):331–339. https://doi.org/10.1007/s11274-008-9897-x
Jo WS, Park HN, Cho DH, Yoo YB, Park SC (2011) Detection of extracellular enzyme activities in Ganoderma neo-japonicum. Mycobiology 39(2):118–120. https://doi.org/10.4489/MYCO.2011.39.2.118
Ijoma GN, Tekere M (2017) Potential microbial applications of co-cultures involving ligninolytic fungi in the bioremediation of recalcitrant xenobiotic compounds. Int J Environ Sci Technol 14(8):1787–1806. https://doi.org/10.1007/s13762-017-1269-3
Kobakhidze A, Asatiani M, Kachlishvili E, Elisashvili V (2016) Induction and catabolite repression of cellulase and xylanase synthesis in the selected white-rot basidiomycetes. Ann Agrar Sci 14(3):169–176. https://doi.org/10.1016/j.aasci.2016.07.001
Ahmadi Khozani M, Emtiazi G, Aghaei SS, Ghasemi SM, Zolfaghari MR. Application of fungal laccase for heavy metals precipitation using tannin as a natural mediator. Int J Environ Sci Technol. 2020;(0123456789). https://doi.org/10.1007/s13762-020-02992-7
Terrasan CRF, Carmona EC (2015) Solid-state fermentation of brewer’s spent grain for xylanolytic enzymes production by Penicilliumjanczewskii and analyses of the fermented substrate. Biosci J 31(6):1826–36. https://doi.org/10.14393/BJ-v31n6a2015-30044
Martinez-Carrera D, Aguilar A, Martinez W, Bonilla M, Morales P, Sobal M. Mushroom cultivation on coffee pulp. Commer Prod Mark Edible Mushrooms Cultiv Coffee Pulp Mex. 2018;471–88.
Lynch KM, Steffen EJ, Arendt EK (2016) Brewers’ spent grain: a review with an emphasis on food and health. J Inst Brew 122(4):553–568. https://doi.org/10.1002/jib.363
Lomascolo A, Uzan-Boukhris E, Sigoillot JC, Fine F (2012) Rapeseed and sunflower meal: a review on biotechnology status and challenges. Appl Microbiol Biotechnol 95(5):1105–1114. https://doi.org/10.1007/s00253-012-4250-6
Murthy PS, Madhava NM (2012) Sustainable management of coffee industry by-products and value addition - a review. Resour Conserv Recycl 66:45–58. https://doi.org/10.1016/j.resconrec.2012.06.005
Wang F, Terry N, Xu L, Zhao L, Ding Z, Ma H. 2019 Fungal laccase production from lignocellulosic agricultural wastes by solid-state fermentation: a review. Vol. 7, Microorganisms. MDPI AG; https://doi.org/10.3390/microorganisms7120665
Milić MD, Buntić AV, Mihajlovski KR, Ilić NV, Davidović SZ, Dimitrijević-Branković SI (2021) The development of a combined enzymatic and microbial fermentation as a viable technology for the spent coffee ground full utilization. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-021-01605-8
Shah TA, Ullah R (2019) Pretreatment of wheat straw with ligninolytic fungi for increased biogas productivity. Int J Environ Sci Technol 16(11):7497–7508. https://doi.org/10.1007/s13762-019-02277-8
Różyło K, Bohacz J (2020) Microbial and enzyme analysis of soil after the agricultural utilization of biogas digestate and mineral mining waste. Int J Environ Sci Technol 17(2):1051–1062. https://doi.org/10.1007/s13762-019-02522-0
Zhu N, Liu J, Yang J, Lin Y, Yang Y, Ji L et al (2016) Comparative analysis of the secretomes of Schizophyllum commune and other wood-decay basidiomycetes during solid-state fermentation reveals its unique lignocellulose-degrading enzyme system. Biotechnol Biofuels 9(1):1–22. https://doi.org/10.1186/s13068-016-0461-x
Seifert KA, Rossman AY (2010) How to describe a new fungal species. IMA Fungus 1(2):109–116
Carrillo-Nieves D, Saldarriaga-Hernandez S, Gutiérrez-Soto G, Rostro-Alanis M, Hernández-Luna C, Alvarez AJ et al (2020) Biotransformation of agro-industrial waste to produce lignocellulolytic enzymes and bioethanol with a zero waste. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-020-00738-6
Mihajlovski K, Radovanović Ž, Carević M, Dimitrijević-Branković S (2018) Valorization of damaged rice grains: optimization of bioethanol production by waste brewer’s yeast using an amylolytic potential from the Paenibacilluschitinolyticus CKS1. Fuel 224(March):591–599. https://doi.org/10.1016/j.fuel.2018.03.135
Nargotra P, Sharma V, Sharma S, Kapoor N, Bajaj BK (2020) Development of consolidated bioprocess for biofuel-ethanol production from ultrasound-assisted deep eutectic solvent pretreated Partheniumhysterophorus biomass. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-020-01017-0
Bušić A, Mardetko N, Kundas S, Morzak G, Belskaya H, Šantek MI et al (2018) Bioethanol production from renewable raw materials and its separation and purification: a review. Food Technol Biotechnol 56(3):289–311. https://doi.org/10.17113/ftb.56.03.18.5546
Zhang YJ, Zhang S, Liu XZ, Wen HA, Wang M (2010) A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Lett Appl Microbiol 51(1):114–118. https://doi.org/10.1111/j.1472-765X.2010.02867.x
Jović J, Buntić A, Radovanović N, Petrović B, Mojović L (2018) Lignin-degrading abilities of novel autochthonous fungal isolates Trameteshirsuta F13 and Stereumgausapatum F28. Food Technol. Biotechnol 56(3):354–365. https://doi.org/10.17113/ftb.56.03.18.5348
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct se-quencing of fungal ribosomal RNA genes for phylogenetics. In: PCR – protocols and applications – a laboratory manual. Cambridge, MA, USA: Academic Press, Inc 1990. pp. 315- 22.
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr Microbiol 57(5):503–507. https://doi.org/10.1007/s00284-008-9276-8
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Mihajlovski K, Buntić A, Milić M, Rajilić-Stojanović M, Dimitrijević-Branković S (2021) From agricultural waste to biofuel: enzymatic potential of a bacterial isolate Streptomyces fulvissimus CKS7 for bioethanol production. Waste Biomass Valori 12(1):165–174. https://doi.org/10.1007/s12649-020-00960-3
Rittenour WR, Ciaccio CE, Barnes CS, Kashon ML, Lemons AR, Beezhold DH et al (2014) Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environ Sci Process Impacts 16(1):33–43. https://doi.org/10.1039/C3EM00441D
Jatt AN, Tunio SA, Memon SB, Qureshi AS, Bhutto MA. API-ZYM enzymatic profile of. 2018;50(3):977–81. https://doi.org/10.17582/journal.pjz/2018.50.3.977.981
Kavkler K, Gunde-Cimerman N, Zalar P, Demšar A (2015) Fungal contamination of textile objects preserved in Slovene museums and religious institutions. Int Biodeterior Biodegrad 97:51–59. https://doi.org/10.1016/j.ibiod.2014.09.020
Hadda M, Djamel C, Akila O (2015) Screening of extracellular enzyme activities of Ganoderma and Fomes species collected from northern east Algeria. Res J Pharm Biol Chem Sci 6(4):1455–1462
Pandey AK, Vishwakarma SK, Srivastava AK, Pandey VK, Agrawal S, Singh MP (2014) Production of ligninolytic enzymes by white rot fungi on lignocellulosic wastes using novel pretreatments. Cell Mol Biol 60(5):41–5. https://doi.org/10.14715/cmb/2014.60.5.8
Sandra M, Laura L (2015) Production of lignocellulolytic enzymes from three white-rot fungi by solid-state fermentation and mathematical modeling. African J Biotechnol 14(15):1304–1317. https://doi.org/10.5897/AJB2014.14331
Ding C, Wang X, Li M (2019) Evaluation of six white-rot fungal pretreatments on corn stover for the production of cellulolytic and ligninolytic enzymes, reducing sugars, and ethanol. Appl Microbiol Biotechnol 103(14):5641–5652. https://doi.org/10.3390/microorganisms7120665
Quiroz-Castañeda RE, Balcázar-López E, Dantán-González E, Martinez A, Folch-Mallol J, Martínez-Anaya C. 2009 Characterization of cellulolytic activities of Bjerkandera adusta and Pycnoporus sanguineus on solid wheat straw medium. Electron J Biotechnol 12(4). https://doi.org/10.2225/vol12- issue4-fulltext-3
Papinutti VL, Diorio LA, Forchiassin F (2003) Production of laccase and manganese peroxidase by Fomessclerodermeus grown on wheat bran. J Ind Microbiol Biotechnol 30(3):157–160. https://doi.org/10.1007/s10295-003-0025-5
Kumar B, Bhardwaj N, Alam A, Agrawal K, Prasad H, Verma P (2018) Production, purification and characterization of an acid/alkali and thermo tolerant cellulase from Schizophyllum commune NAIMCC-F-03379 and its application in hydrolysis of lignocellulosic wastes. AMB Express 8(1):173. https://doi.org/10.1186/s13568-018-0696-y
Quiroz-Castañeda RE, Pérez-Mejía N, Martínez-Anaya C, Acosta-Urdapilleta L, Folch-Mallol J (2011) Evaluation of different lignocellulosic substrates for the production of cellulases and xylanases by the basidiomycete fungi Bjerkanderaadusta and Pycnoporussanguineus. Biodegradation 22(3):565–72. https://doi.org/10.1007/s10532-010-9428-y
Haq IU, Ashraf H, Iqbal J, Qadeer MA (2003) Production of alpha amylase by Bacillus licheniformis using an economical medium. Bioresour Technol 87(1):57–61
Rajoka MI, Huma T, Khalid AM, Latif F (2005) Kinetics of enhanced substrate consumption and endo-β-xylanase production by a mutant derivative of Humicolalanuginosa in solid-state fermentation. World J Microbiol Biotechnol 21(6–7):869–876. https://doi.org/10.1007/s11274-004-6030-7
Erdem E, Ucar MC, Kaymaz Y, Pazarlioglu NK (2009) New and different lignocellulosic materials from Turkey for laccase and manganese peroxidase production by Trametes versicolor. Eng Life Sci 9(1):60–65. https://doi.org/10.1002/elsc.200700025
Fan L, Pandey A, Mohan R, Soccol CR (2000) Use of various coffee industry residues for the cultivation of Pleurotusostreatus in solid state fermentation. Acta Biotechnol 20(1):41–52. https://doi.org/10.1002/abio.370200108
Tripathi A, Upadhyay RC, Singh S (2012) Extracellular ligninolytic enzymes in Bjerkanderaadusta and Lentinussquarrosulus. Indian J Microbiol 52(3):381–387. https://doi.org/10.1007/s12088-011-0232-0
Belcarz A, Ginalska G, Kornillowicz-Kowalska T (2005) Extracellular enzyme activities of Bjerkanderaadusta R59 soil strain, capable of daunomycin and humic acids degradation. Appl Microbiol Biotechnol 68(5):686–694. https://doi.org/10.1007/s00253-005-1918-1
Kaal EEJ, Field JA, Joyce TW (1995) Increasing ligninolytic enzyme activities in several white-rot Basidiomycetes by nitrogen-sufficient media. Bioresour Technol 53(2):133–139. https://doi.org/10.1016/0960-8524(95)00066-N
Agamuthu, P,A. Nithiya: Waste to enzymes through solid state fermentation. ISWA/APESB World Congress, 12–15 Okt 2009, Lisbon, Portugal. (2009).
Irshad M, Asgher M (2011) Production and optimization of ligninolytic enzymes by white rot fungus Schizophyllum commune IBL-06 in solid state medium banana stalks. African J Biotechnol 10(79):18234–18242. https://doi.org/10.5897/AJB11.2242
Shimazaki M, Matsuki T, Yamauchi K, Iwata M, Takahashi H, Sakamoto K et al (2008) Clinical performance of a salivary amylase activity monitor during hemodialysis treatment. Biomark Insights 2008(3):429–434
Babu CR, Harsha K, Sheik KB, Viswanatha CK (2018) Wheat bran-composition and nutritional quality: a review. Adv Biotechnol Microbiol 9(1):1–7. https://doi.org/10.19080/AIBM.2018.09.555754
Malathi V, Devegowda G (2001) In vitro evaluation of nonstarch polysaccharide digestibility of feed ingredients by enzymes. Poult Sci 80(3):302–305. https://doi.org/10.1093/ps/80.3.302
Ganbarov KG, Kulieva NA, Muradov PZ (2001) Biosynthesis of pectinase by fungi of the genera Bjerkandera and Coriolus during solid-phase fermentation. Appl Biochem Microbiol 37(6):593–595. https://doi.org/10.1023/A:1012303101102
Mehmood T, Saman T, Irfan M, Anwar F, Ikram MS, Tabassam Q (2019) Pectinase production from Schizophyllum commune through central composite design using citrus waste and its immobilization for industrial exploitation. Waste Biomass Valori 10(9):2527–2536. https://doi.org/10.1007/s12649-018-0279-9
Gautam A, Kumar A, Bharti AK, Dutt D (2018) Rice straw fermentation by Schizophyllum commune ARC-11 to produce high level of xylanase for its application in pre-bleaching. J Genet Eng Biotechnol 16(2):693–701. https://doi.org/10.1016/j.jgeb.2018.02.006
Barzee TJ, El- Mashad HM, Zhang R, Pan Z. Carrots. 2019 Integrated processing technologies for food and agricultural by-products. Elsevier Inc. 297–330. https://doi.org/10.1016/B978-0-12-814138-0.00012-5
Lu XB, Zhang YM, Yang J, Liang Y (2007) Enzymatic hydrolysis of corn stover after pretreatment with dilute sulfuric acid. Chem Eng Technol 30(7):938–944. https://doi.org/10.1002/ceat.200700035
Li Y, Ruan R, Chen PL, Liu Z, Pan X, Lin X et al (2004) Enzymatic hydrolysis of corn stover pretreated by combined dilute alkaline treatment and homogenization. Trans Am Soc Agric Eng 47(3):821–5. https://doi.org/10.13031/2013.16078
Rochelle L, Sciences P, Curie BM, Crépeau AM, Rochelle L. 2008 A thermomechanical process for improving enzymatic hydrolysis of brewer ’ s spent grain
Ravindran R, Jaiswal S, Abu-Ghannam N, Jaiswal AK (May 2018) A comparative analysis of pretreatment strategies on the properties and hydrolysis of brewers’ spent grain. Bioresour Technol 2018(248):272–279. https://doi.org/10.1016/j.biortech.2017.06.039
White JS, Yohannan BK, Walker GM (2008) Bioconversion of brewer’s spent grains to bioethanol. FEMS Yeast Res 8(7):1175–1184. https://doi.org/10.1111/j.1567-1364.2008.00390.x
Liguori R, Soccol CR, de Souza Vandenberghe LP, Woiciechowski AL, Faraco V (2015) Second generation ethanol production from brewers’ spent grain. Energies 8(4):2575–2586
Wilkinson S, Smart KA, Cook DJ (2014) Optimisation of alkaline reagent based chemical pre-treatment of Brewers spent grains for bioethanol production. Ind Crops Prod 62:219–227. https://doi.org/10.1016/j.indcrop.2014.08.036
Yohannan BK, White JS, Bennett J, Walker GM. Distiller’s spent grains: a substrate for bioethanol? Distill Spirits. 2012;(May):147–54.
Mahapatra S, Manian R (2020) Enhancement, production, and immobilization of beta-glucosidase from Zobellelladenitrificans VIT SB117 and its utilization in bioethanol production from lignocellulosic feedstock. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-020-00718-w
Wan C, Li Y (2010) Microbial pretreatment of corn stover with Ceriporiopsissubvermispora for enzymatic hydrolysis and ethanol production. Bioresour Technol 101(16):6398–6403. https://doi.org/10.1016/j.biortech.2010.03.070
Acknowledgements
The authors would like to thank agricultural cooperative “Mrkšićevi salaši” for obtaining corn waste.
Funding
This work was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Contract No. 451–03-68/2020–14/200135 and 451–03-9/2021–14/200287) and Bilateral Project Serbia/Croatia (337–00-205/2019–09/35) (2019–2021). Ministarstvo Prosvete,Nauke i Tehnološkog Razvoja,451–03-68/2020–14/200135,451–03-9/2021–14/200287,Bilateral Project Serbia/Croatia (337–00-205/2019–09/35) (2019–2021)
Author information
Authors and Affiliations
Contributions
N. Ilić: planning of study, investigations on isolation of fungi and determination of qualitative and quantitative enzymes activities, data analysis, results discussion, manuscript writing. S. Davidović: investigations, manuscript writing, performing statistics. M. Milić: investigations on fermentation processes, data analysis, results discussion, manuscript writing. M. Rajilić-Stojanović: molecular identification of fungal isolates, editing. Danijela Pecarski: investigations on fermentation processes. Mirela Ivančić-Šantek: investigations on bioethanol production. K. Mihajlovski: planning of study, investigations on bioethanol production, data analysis, results verification, manuscript writing. S. Dimitrijević-Branković: conceptualization, planning, and supervision.
Corresponding author
Ethics declarations
Ethical Approval
All authors agree with the contents of the manuscript and its submission to Biomass Conversion and Biorefinery. This research has not been published before and is not under consideration for publication anywhere.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ilić, N., Davidović, S., Milić, M. et al. Valorization of lignocellulosic wastes for extracellular enzyme production by novel Basidiomycetes: screening, hydrolysis, and bioethanol production. Biomass Conv. Bioref. 13, 17175–17186 (2023). https://doi.org/10.1007/s13399-021-02145-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-021-02145-x