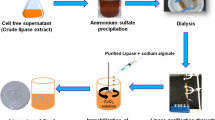
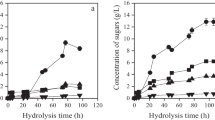
Abstract
Two bacterial and six yeast strains capable of lipase production were investigated for their extracellular lipase (ECL), cell-bound lipase (CBL), and growth profile in basal standard medium (BSM) and the low-cost palm oil mill effluent (POME) media. The yeast Magnusiomyces spicifer AW2 and bacteria Staphylococcus hominis AUP19 were selected and further employed to explore their survivability, lipase activity, and cell biomass production in various POME concentrations. It was revealed that diluted POME 1:1 at pH 7.0 was suitable for yeast and bacterial performance. Furthermore, the development of co-culture between yeast and bacteria was performed by mixing them at a ratio of 1:1. The synthetic co-culture system was successful in improving CBL activity to 3860 U/L. The bioremediation parameter, i.e., oil and grease (O&G) removal, was efficiently achieved 1.1-fold higher than yeast alone and bacterial monoculture at 80.1 ± 1.3%. At the same time, COD removal was highest with the co-culture treatment of 75.9 ± 2.8%, indicating 1.3-fold more effective than yeast monoculture and 1.4-fold better than single bacterial form. The mixed cells of two strains effectively improved biodiesel production from oleic acid of 73.5 ± 3.3% and palm oil of 82.5 ± 0.3% under solvent-free conditions for 72-h reactions at room temperature (30 ± 2 °C), indicating a promising prospect of a combined approach of POME treatment and valorization.
Graphical abstract






Similar content being viewed by others
Code availability
IBM SPSS Statistical Subscription.
Abbreviations
- BSM:
-
Basal standard medium
- EBSM:
-
Enrichment basal standard medium
- CBL:
-
Cell-bound lipase
- CBLs:
-
Cell-bound lipases
- CBM:
-
Cell biomass
- CG:
-
Cell growth
- COD:
-
Chemical oxygen demand
- ECL:
-
Extracellular lipase
- FAME:
-
Fatty acid methyl ester
- NA:
-
Nutrient agar
- NB:
-
Nutrient broth
- O&G:
-
Oil and grease
- POME:
-
Palm oil mill effluent
- RBM:
-
Rhodamine B medium
- TKN:
-
Total Kjeldahl Nitrogen
- YM:
-
Yeast malt
- YMA:
-
Yeast malt agar
- YMB:
-
Yeast malt broth
References
Basheer SM, Chellappan S, Beena PS, Sukumaran RK, Elyas KK, Chandrasekaran M (2011) Lipase from marine Aspergillus awamori BTMFW032: production, partial purification and application in oil effluent treatment. N Biotechnol. 28(6):627–38. https://doi.org/10.1016/j.nbt.2011.04.007
Bornscheuer UT (2018) Enzymes in lipid modification. Annu Rev Food Sci Technol 9:85–103
Amini Z, Ilham Z, Ong HC, Mazaheri H, Chen W-H (2017) State of the art and prospective of lipase-catalyzed transesterification reaction for biodiesel production. Energy Convers Manag 141:339–353. https://doi.org/10.1016/j.enconman.2016.09.049
López EN, Medina AR, Moreno PAG, Cerdán LE, Valverde LM, Grima EM (2016) Biodiesel production from Nannochloropsis gaditana lipids through transesterification catalyzed by Rhizopus oryzae lipase. Bioresour Technol 203:236–244
Canet A, Bonet-Ragel K, Benaiges MD, Valero F (2016) Lipase-catalysed transesterification: viewpoint of the mechanism and influence of free fatty acids. Biomass Bioenergy 85:94–99
Amini Z, Ong HC, Harrison MD, Kusumo F, Mazaheri H, Ilham Z (2017) Biodiesel production by lipase-catalyzed transesterification of Ocimum basilicum L. (sweet basil) seed oil. Energy Convers Manag 132:82–90
Joshi R, Kuila A (2018) Lipase and their different industrial applications: a review. Brazilian J Biol Sci 5(10):237–47. https://doi.org/10.21472/bjbs.051004
Melani NB, Tambourgi EB, & Silveira E (2019). Lipases: from production to applications. Sep Purif Rev. 1–16. https://doi.org/10.1080/15422119.2018.1564328
Louhasakul Y, Cheirsilp B, Prasertsan P (2016) Valorization of palm oil mill effluent into lipid and cell-bound lipase by marine yeast Yarrowia lipolytica and their application in biodiesel production. Waste Biomass Valori 7(3):417–426. https://doi.org/10.1007/s12649-015-9451-7
Kumar S, Katiyar N, Ingle P, Negi S (2011) Use of evolutionary operation (EVOP) factorial design technique to develop a bioprocess using grease waste as a substrate for lipase production. Bioresour Technol 102(7):4909–4912
Kumar S, Mathur A, Singh V, Nandy S, Khare SK, Negi S (2012) Bioremediation of waste cooking oil using a novel lipase produced by Penicillium chrysogenum SNP5 grown in solid medium containing waste grease. Bioresour Technol 120:300–304
Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl DA (2007). Metabolic modeling of a mutualistic microbial community. Mol Syst Biol 3(92):1–14. https://doi.org/10.1038/msb4100131
Arous F, Hamdi C, Kmiha S, Khammassi N, Ayari A, Neifar M, Mechichi T, Jaouani A (2018) Treatment of olive mill wastewater through employing sequencing batch reactor: performance and microbial diversity assessment. 3 Biotech 8(11):481. https://doi.org/10.1007/s13205-018-1486-6
Shong J, Jimenez Diaz MR, Collins CH (2012) Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol 23(5):798–802. https://doi.org/10.1016/j.copbio.2012.02.001
Zhang C, Qi J, Cao Y (2014) Synergistic effect of yeast-bacterial co-culture on bioremediation of oil-contaminated soil. Bioremediat J. https://doi.org/10.1080/10889868.2013.847402
Ungcharoenwiwat P, H-Kittikun A (2015) Purification and characterization of lipase from Burkholderia sp. EQ3 isolated from wastewater from a canned fish factory and its application for the synthesis of wax esters. J Mol Catal B Enzym 115:96–104. https://doi.org/10.1016/j.molcatb.2015.02.005
Soleimaninanadegani M, Manshad S (2014) Enhancement of biodegradation of palm oil mill effluents by local isolated microorganisms. Int Sch Res Not. 2014:727049. https://doi.org/10.1155/2014/727049
Yele VU, Desai K (2015) A new thermostable and organic solvent-tolerant lipase from Staphylococcus warneri; optimization of media and production conditions using statistical methods. Appl Microbiol Biotechnol 175(2):855–69. https://doi.org/10.1007/s12010-014-1331-2
Wang D, Xu Y, Teng Y (2007) Synthetic activity enhancement of membrane-bound lipase from Rhizopus chinensis by pretreatment with isooctane. Bioprocess Biosyst Eng 30(3):147–155
Vakhlu J, Kour A (2006) Yeast lipases: enzyme purification, biochemical properties and gene cloning. Electron J Biotechnol 9(1):69–85. https://doi.org/10.2225/vol9-issue1-fulltext-9
De Almeida AF, Tauk-Tornisielo SM, Carmona EC (2013). Acid lipase from Candida viswanathii: production, biochemical properties, and potential application. Biomed Res Int. https://doi.org/10.1155/2013/435818
Baloch KA, Upaichit A, Cheirsilp B, Fibriana F (2021) The occurrence of triple catalytic characteristics of yeast lipases and their application prospects in biodiesel production from non-edible Jatropha curcas oil in a solvent-free system. Curr Microbiol. https://doi.org/10.1007/s00284-021-02448-2
Srimhan P, Kongnum K, Taweerodjanakarn S, Hongpattarakere T (2011) Selection of lipase producing yeasts for methanol-tolerant biocatalyst as whole cell application for palm-oil transesterification. Enzym Microb Technol 48(3):293–8. https://doi.org/10.1016/j.enzmictec.2010.12.004
de Carvalho CCCR (2017) Whole cell biocatalysts: essential workers from nature to the industry. Microb Biotechnol 10(2):250–263
Ladkau N, Schmid A, Bühler B (2014) The microbial cell—functional unit for energy dependent multistep biocatalysis. Curr Opin Biotechnol 30:178–189
Lin B, Tao Y (2017) Whole-cell biocatalysts by design. Microb Cell Fact 16(1):106. https://doi.org/10.1186/s12934-017-0724-7
Yan J, Yan Y, Madzak C, Han B (2017) Harnessing biodiesel-producing microbes: from genetic engineering of lipase to metabolic engineering of fatty acid biosynthetic pathway. Crit Rev Biotechnol 37(1):26–36
Rosenstein R, & Götz F (2000). Staphylococcal lipases: biochemical and molecular characterization. Biochimie. https://doi.org/10.1016/S0300-9084(00)01180-9
Hasan F, Shah AA, Hameed A (2006) Influence of culture conditions on lipase production by Bacillus sp. FH5. Ann Microbiol. 56(3):247–52
Sooch BS, Kauldhar BS (2013). Influence of multiple bioprocess parameters on production of lipase from Pseudomonas sp. BWS-5. Brazilian Arch Biol Technol. https://doi.org/10.1590/S1516-89132013000500002
Behera AR, Veluppal A, Dutta K (2019) Optimization of physical parameters for enhanced production of lipase from Staphylococcus hominis using response surface methodology. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-019-04304-0
Hasan-Beikdashti H, Safiarian MS, Amerib A, Ghahremanic MH, Khoshayandd M, R;Faramarzi, M A MF, (2012) Optimization of culture conditions for production of lipase by a newly isolated bacterium Stenotrophomonas maltophilia. J Taiwan Inst Chem Eng 43:670–677. https://doi.org/10.1016/j.jtice.2012.03.005
Ahmad AL, Chan CY, Abd Shukor SR, Mashitah MD (2008) Recovery of oil and carotenes from palm oil mill effluent (POME). Chem Eng J 141(1–3):383–386. https://doi.org/10.1016/j.cej.2008.03.005
Primandari SRP, Yaakob Z, Mohammad M, Mohamad AB (2013) Characteristics of residual oil extracted from palm oil mill effluent (POME). World Appl Sci J 27(11):1482–1484
Matinja AI, Mohd Zain NA, Suhaimi MS, Alhassan AJ (2019) Optimization of biodiesel production from palm oil mill effluent using lipase immobilized in PVA-alginate-sulfate beads. Renew Energy 135:1178–1185. https://doi.org/10.1016/j.renene.2018.12.079
Cheng J, Zhu X, Ni J, Borthwick A (2010) Palm oil mill effluent treatment using a two-stage microbial fuel cells system integrated with immobilized biological aerated filters. Bioresour Technol 101(8):2729–2734
Poh PE, Yong W-J, Chong MF (2010) Palm oil mill effluent (POME) characteristic in high crop season and the applicability of high-rate anaerobic bioreactors for the treatment of POME. Ind Eng Chem Res 49(22):11732–11740. https://doi.org/10.1021/ie101486w
Salihu A, Alam MZ, AbdulKarim MI, Salleh HM (2011) Optimization of lipase production by Candida cylindracea in palm oil mill effluent based medium using statistical experimental design. J Mol Catal B Enzym 69(1–2):66–73. https://doi.org/10.1016/j.molcatb.2010.12.012
Bleve G, Lezzi C, Chiriatti MA, D’Ostuni I, Tristezza M, Di Venere D, Sergio L, Mita G, Grieco F (2011) Selection of non-conventional yeasts and their use in immobilized form for the bioremediation of olive oil mill wastewaters. Bioresour Technol. 102(2):982–9. https://doi.org/10.1016/j.biortech.2010.09.059
Islam MA, Yousuf A, Karim A, Pirossi D, Khan MR, Wahid ZA (2018) Bioremediation of palm oil mill effluent and lipid production by Lipomyces starkeyi: a combined approach. J Clean Prod 172:1779–1787. https://doi.org/10.1016/j.jclepro.2017.12.012
Jallouli R, Bezzine S (2015) Lipase production by a Tunisian Fusarium solani strain cultivated on olive oil wastewater-based media and a biotreatment assay. Desalin Water Treat 57(43):20327–20331. https://doi.org/10.1080/19443994.2015.1111809
Papanikolaou S, Dimou A, Fakas S, Diamantopoulou P, Philippoussis A, Galiotou-Panayotou M, Aggelis G (2011). Biotechnological conversion of waste cooking olive oil into lipid-rich biomass using Aspergillus and Penicillium strains. J Appl Microbiol. https://doi.org/10.1111/j.1365-2672.2011.04961.x
Theerachat M, Tanapong P, Chulalaksananukul W (2017) The culture or co-culture of Candida rugosa and Yarrowia lipolytica strain rM-4A, or incubation with their crude extracellular lipase and laccase preparations, for the biodegradation of palm oil mill wastewater. Int Biodeterior Biodegradation 121:11–18
Karim A, Islam MA, Mishra P, Muzahid AJM, Yousuf A, Khan MMR, Faizal CKM (2021). Yeast and bacteria co-culture-based lipid production through bioremediation of palm oil mill effluent: a statistical optimization. Biomass Conver Bior. https://doi.org/10.1007/s13399-021-01275-6
Pai A, Tanouchi Y, Collins CH, You L (2009) Engineering multicellular systems by cell–cell communication. Curr Opin Biotechnol 20(4):461–470
Jawed K, Yazdani SS, Koffas MA (2019) Advances in the development and application of microbial consortia for metabolic engineering. Metab Eng Commun. 9:e00095. https://doi.org/10.1016/j.mec.2019.e00095
Song H, Ding M-Z, Jia X-Q, Ma Q, Yuan Y-J (2014) Synthetic microbial consortia: from systematic analysis to construction and applications. Chem Soc Rev 43:6954. https://doi.org/10.1039/C4CS00114A10.1039/c4cs00114a
Fleet GH (1990) Growth of yeasts during wine fermentations. J Wine Res 1(3):211–223
Guilloux-Benatier M, Remize F, Gal L, Guzzo J, Alexandre H (2006) Effects of yeast proteolytic activity on Oenococcus oeni and malolactic fermentation. FEMS Microbiol Lett 263(2):183–188
Vasserot Y, Caillet S, Maujean A (1997) Study of anthocyanin adsorption by yeast lees. Effect of some physicochemical parameters. Am J Enol Vitic. 48(4):433–7
Karim A, Islam MA, Yousuf A, Khan MMR, Faizal CKM (2019) Microbial lipid accumulation through bioremediation of palm oil mill wastewater by Bacillus cereus. ACS Sustain Chem Eng 7(17):14500–14508
Karim A, Islam MA, Bin KZ, Yousuf A, Khan MR, Faizal CKM (2021) Microbial lipid accumulation through bioremediation of palm oil mill effluent using a yeast-bacteria co-culture. Renew Energy 176:106–114. https://doi.org/10.1016/j.renene.2021.05.055
Rachmadona N, Quayson E, Amoah J, Alfaro-Sayes DA, Hama S, Aznury M, Kondo A, Ogino C (2021) Utilizing palm oil mill effluent (POME) for the immobilization of Aspergillus oryzae whole-cell lipase strains for biodiesel synthesis. Biofuels, Bioprod Biorefining 15(3):804–814
Atik Mas-Ud M, Roushan Ali M, Zia Hasan SM, Ashikul Islam M, Faruk Hasan M, Asadul Islam M, Sikdar B (2020). Molecular detection and biological control of human hair dandruff causing microorganism Staphylococcus aureus. J Pure Appl Microbiol. https://doi.org/10.22207/JPAM.14.1.16
Wang L, Chi Z, Wang X, Liu Z, Li J (2007) Diversity of lipase-producing yeasts from marine environments and oil hydrolysis by their crude enzymes. Ann Microbiol 57(4):495–501
Ban K, Kaieda M, Matsumoto T, Kondo A, Fukuda H (2001) Whole cell biocatalyst for biodiesel fuel production utilizing Rhizopus oryzae cells immobilized within biomass support particles. Biochem Eng J 8(1):39–43. https://doi.org/10.1016/S1369-703x(00)00133-9
Yan J, Yan Y (2008) Optimization for producing cell-bound lipase from Geotrichum sp. and synthesis of methyl oleate in microaqueous solvent. Appl Microbiol Biotechnol 78(3):431–9
Adachi D, Koh F, Hama S, Ogino C, Kondo A (2013) A robust whole-cell biocatalyst that introduces a thermo- and solvent-tolerant lipase into Aspergillus oryzae cells: characterization and application to enzymatic biodiesel production. Enzym Microb Technol 52(6–7):331–5. https://doi.org/10.1016/j.enzmictec.2013.03.005
Adachi D, Hama S, Nakashima K, Bogaki T, Ogino C, Kondo A (2013) Production of biodiesel from plant oil hydrolysates using an Aspergillus oryzae whole-cell biocatalyst highly expressing Candida antarctica lipase B. Bioresour Technol 135:410–416. https://doi.org/10.1016/j.biortech.2012.06.092
Nuylert A, Hongpattarakere T (2012) Improvement of cell-bound lipase from Rhodotorula mucilaginosa P11I89 for use as a methanol-tolerant, whole-cell biocatalyst for production of palm-oil biodiesel. Ann Microbiol 63(3):929–939. https://doi.org/10.1007/s13213-012-0546-0
Oh H, Lee HJ, Lee J, Jo C, Yoon Y (2019) Identification of microorganisms associated with the quality improvement of dry-aged beef through microbiome analysis and DNA sequencing, and evaluation of their effects on beef quality. J Food Sci 84(10):2944–2954
Rice EW, Baird RB, Eaton AD, Clesceri LS (2012) Standard methods for the examination of water and wastewater. Am Public Heal Assoc Washington, DC, p 541
Cheirsilp B, Suwannarat W, Niyomdecha R (2011) Mixed culture of oleaginous yeast Rhodotorula glutinis and microalga Chlorella vulgaris for lipid production from industrial wastes and its use as biodiesel feedstock. N Biotechnol 28(4):362–368
Botthoulath V, Upaichit A, Thumarat U (2018) Identification and in vitro assessment of potential probiotic characteristics and antibacterial effects of Lactobacillus plantarum subsp. plantarum SKI19, a bacteriocinogenic strain isolated from Thai fermented pork sausage. J Food Sci Technol 55(7):2774–85
Kwon DY, Rhee JS (1986) A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J Am Oil Chem Soc 63:89–92. https://doi.org/10.1007/BF02676129
Lee SY, Rhee JS (1994) Hydrolysis of triglyceride by the whole cell of Pseudomonas putida 3SK in two-phase batch and continuous reactor systems. Biotechnol Bioeng 44:437–443
Baloch KA, Upaichit A, Cheirsilp B (2019) Use of low-cost substrates for cost-effective production of extracellular and cell-bound lipases by a newly isolated yeast Dipodascus capitatus A4C. Biocatal Agric Biotechnol 19:101012. https://doi.org/10.1016/j.bcab.2019.101102
Prasertsan P, Binmaeil H (2018) Treatment of oil mill effluent by thermotolerant polymer-producing fungi. J Water Environ Technol 16(3):127–137. https://doi.org/10.2965/jwet.17-031
Bala LJ, Ismail N (2014) Biodegradation of palm oil mill effluent (POME) by bacterial. Int J Sci Res Publ 4(3):1–10
Nur MMA, Swaminathan MK, Boelen P, Buma AGJ (2019) Sulfated exopolysaccharide production and nutrient removal by the marine diatom Phaeodactylum tricornutum growing on palm oil mill effluent. J Appl Phycol 31(4):2335–2348
Ryu S, Karim MN (2011) A whole cell biocatalyst for cellulosic ethanol production from dilute acid-pretreated corn stover hydrolyzates. Appl Microbiol Biotechnol. 91(3):529–42. https://doi.org/10.1007/s00253-011-3261-z
Kamyab H, Chelliapan S, Din MFM, Rezania S, Khademi T, Kumar A (2018) Palm oil mill effluent as an environmental pollutant. Palm oil 13:13–28
Mamimin C, Thongdumyu P, Hniman A, Prasertsan P, Imai T, O-Thong S (2012). Simultaneous thermophilic hydrogen production and phenol removal from palm oil mill effluent by Thermoanaerobacterium-rich sludge. In: International Journal of Hydrogen Energy. Pergamon. https://doi.org/10.1016/j.ijhydene.2012.04.062
Ilesanmi OI, Adekunle AE, Omolaiye JA, Olorode EM, Ogunkanmi AL (2020) Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Sci African. https://doi.org/10.1016/j.sciaf.2020.e00279
Rattana T (2011). Isolation of proteolytic, lipolytic, and bioemulsifying bacteria for improvement of the aerobic treatment of poultry processing wastewater. African J Microbiol Res. https://doi.org/10.5897/ajmr11.824
Esakkiraj P, Austin Jeba Dhas G, Palavesam A, Immanuel G (2010) Media preparation using tuna-processing wastes for improved lipase production by shrimp gut isolate Staphylococcus epidermidis CMST Pi 2. Appl Microbiol Biotechnol. 160(4):1254–65. https://doi.org/10.1007/s12010-009-8632-x
Jarvis GN, Thiele JH (1997) Qualitative Rhodamine B assay which uses tallow as a substrate for lipolytic obligately anaerobic bacteria. J Microbiol Methods. https://doi.org/10.1016/S0167-7012(97)00991-3
Wang Y, Srivastava KC, Shen GJ, Wang HY (1995) Thermostable alkaline lipase from a newly isolated thermophilic Bacillus, strain A30–1 (ATCC 53841). J Ferment Bioeng. https://doi.org/10.1016/0922-338X(95)91257-6
Sandi J, Mata-Araya I, & Aguilar F (2020). Diversity of lipase-producing microorganisms from tropical oilseeds Elaeis guineensis, Ricinus communis, and Jatropha curcas L. from Costa Rica. Curr Microbiol. https://doi.org/10.1007/s00284-020-01886-8
Marimuthu K (2013) Isolation and characterization of Staphylococcus hominis JX961712 from oil contaminated soil. J Pharm Res. https://doi.org/10.1016/j.jopr.2013.03.014
Joseph B, Ramteke PW, Kumar PA (2006) Studies on the enhanced production of extracellular lipase by Staphylococcus epidermidis. J Gen Appl Microbiol 52(6):315–320
Edupuganti S, Parcha L, Narasu Mangamoori L (2017) Purification and characterization of extracellular lipase from Staphylococcus epidermidis (MTCC 10656). J Appl Pharm Sci 7(01):57–063. https://doi.org/10.7324/JAPS.2017.70108
Paul A, Stösser TR, Zehl A, Zwirnmann E, Vogt RD, Steinberg CEW (2006) Nature and abundance of organic radicals in natural organic matter: effect of pH and irradiation. Environ Sci Technol 40(19):5897–5903. https://doi.org/10.1021/es060742d
Fernández-Calviño D, Bååth E (2010) Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol Ecol 73(1):149–156. https://doi.org/10.1111/j.1574-6941.2010.00873.x
Kennedy MB, Lennarzg WJ (1979) Characterization of the extracellular lipase of Bacillus subtilis and its relationship to a membrane-bound lipase found in a mutant strain. J Biol Chem 254(4):1080–1089. https://doi.org/10.1016/S0021-9258(17)34170-4
Sharma R, Chisti Y, Banerjee UC (2001) Production, purification, characterization, and applications of lipases. Biotechnol Adv 19(8):627–662
Hlavsová K, Zarevúcka M, Wimmer Z, Macková M, Sovová H (2009) Geotrichum candidum 4013: extracellular lipase versus cell-bound lipase from the single strain. J Mol Catal B Enzym 61(3–4):188–193. https://doi.org/10.1016/j.molcatb.2009.06.012
Salgado V, Fonseca C, Lopes da Silva T, Roseiro JC, Eusébio A (2019) Isolation and identification of Magnusiomyces capitatus as a lipase-producing yeast from olive mill wastewater. Waste Biomass Valori. https://doi.org/10.1007/s12649-019-00725-7
Salgado JM, Abrunhosa L, Venâncio A, Domínguez JM, Belo I (2016) Combined bioremediation and enzyme production by Aspergillus sp. in olive mill and winery wastewaters. Int Biodeterior Biodegrad 110:16–23. https://doi.org/10.1016/j.ibiod.2015.12.011
Edwards VH (1970) The influence of high substrate concentrations on microbial kinetics. Biotechnol Bioeng 12(5):679–712
Liu S (2020) Bioprocess engineering: kinetics, sustainability, and reactor design. Elsevier. https://doi.org/10.1016/B978-0-12-821012-3.00021-X
Habib MAB, Yusoff FM, Phang SM, Ang KJ, Mohamed S (1997) Nutritional values of chironomid larvae grown in palm oil mill effluent and algal culture. Aquaculture 158(1–2):95–105
Yousuf A, Sannino F, Addorisio V, Pirozzi D (2010) Microbial conversion of olive oil mill wastewaters into lipids suitable for biodiesel production. J Agric Food Chem 58(15):8630–8635. https://doi.org/10.1021/jf101282t
Saidu H, Salau OA, Mohamad SE. Investigating the effect of several palm oil mill effluent (POME) dilutions on biomass and specific growth rate of C. sorokiniana. Int J Life Sci Biotechnol 4(2):174–185. https://doi.org/10.38001/ijlsb.827309
Sabra W, & Zeng A-P (2014). Mixed microbial cultures for industrial biotechnology: success, chance, and challenges. Grunwald P, editor. Pan Stanford Publishing Pte.Ltd.; 2014.
Kunisawa J, Kiyono H (2011) Peaceful mutualism in the gut: revealing key commensal bacteria for the creation and maintenance of immunological homeostasis. Cell Host Microbe 9:83–84
Thompson DS, Carlisle PL, & Kadosh D (2011). Coevolution of morphology and virulence in Candida species [Internet]. Vol. 10, Eukaryotic Cell. Eukaryot Cell; 2011 [cited 2021 Feb 24]. p. 1173–82. https://doi.org/10.1128/EC.05085-11
Steffan BN, Venkatesh N, Keller NP (2020) Let’s get physical: bacterial-fungal interactions and their consequences in agriculture and health. J Fungi 6(4):243
Brenner K, You L, Arnold FH (2008) Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol 26(9):483–489
Bhatia SK, Yi D-H, Kim Y-H, Kim H-J, Seo H-M, Lee J-H, Kim J-H, Jeon J-M, Jang K-S, Kim Y-G, Yang Y-H (2015) Development of semi-synthetic microbial consortia of Streptomyces coelicolor for increased production of biodiesel (fatty acid methyl esters). Fuel 159:189–196. https://doi.org/10.1016/j.fuel.2015.06.084
Lanciotti R, Gianotti A, Baldi D, Angrisani R, Suzzi G, Mastrocola D, Guerzoni ME (2005) Use of Yarrowia lipolytica strains for the treatment of olive mill wastewater. Bioresour Technol 96(3):317–22. https://doi.org/10.1016/j.biortech.2004.04.009
Rodriguez-Mateus Z, Agualimpia B, Zafra G (2016) Isolation and molecular characterization of microorganisms with potential for the degradation of oil and grease from palm oil refinery wastes. Chem Eng Trans 49:517–522. https://doi.org/10.3303/cet1649087
Sung C, Kim BG, Kim S, Joo HS, Kim PI (2010) Probiotic potential of Staphylococcus hominis MBBL 2–9 as anti-Staphylococcus aureus agent isolated from the vaginal microbiota of a healthy woman. J Appl Microbiol 108(3):908–916. https://doi.org/10.1111/j.1365-2672.2009.04485.x
Hwang H, Lee HJ, Lee MA, Sohn H, Chang YH, Han SG, Jeong JY, Lee SH, Hong SW (2020) Selection and characterization of Staphylococcus hominis subsp. hominis wikim0113 isolated from kimchi as a starter culture for the production of natural pre-converted nitrite. Food Sci Anim Resour. 40(4):512–26. https://doi.org/10.5851/kosfa.2020.e29
Marsidi N (2013) Biological treatment of POME (high acidic content): factorial analysis [Bachelor Degree Thesis, Universiti Malaysia Pahang]. UMP Repository. http://umpir.ump.edu.my
Geoffry K, Achur RN (2017) A novel halophilic extracellular lipase with both hydrolytic and synthetic activities. Biocatal Agric Biotechnol 12:125–130. https://doi.org/10.1016/j.bcab.2017.09.012
Almyasheva NR, Shuktueva MI, Petrova DA, Kopitsyn DS, Kotelev MS, Vinokurov VA, Novikov AA (2018) Biodiesel fuel production by Aspergillus niger whole-cell biocatalyst in optimized medium. Mycoscience 59(2):147–152. https://doi.org/10.1016/j.myc.2017.09.003
Chen G, Liu J, Qi Y, Yao J, Yan B (2016) Biodiesel production using magnetic whole-cell biocatalysts by immobilization of Pseudomonas mendocina on Fe3O4-chitosan microspheres. Biochem Eng J 113:86–92. https://doi.org/10.1016/j.bej.2016.06.003
Nain P, Jaiswal SK, Prakash NT, Prakash R, Gupta SK (2020) Influence of acyl acceptor blends on the ester yield and fuel properties of biodiesel generated by whole-cell catalysis of cottonseed oil. Fuel 259:116258. https://doi.org/10.1016/j.fuel.2019.116258
Galeano JD, Mitchell DA, Krieger N (2017) Biodiesel production by solvent-free ethanolysis of palm oil catalyzed by fermented solids containing lipases of Burkholderia contaminans. Biochem Eng J 127:77–86. https://doi.org/10.1016/j.bej.2017.08.008
Tanaka T, Yamada R, Ogino C, Kondo A (2012) Recent developments in yeast cell surface display toward extended applications in biotechnology. Appl Microbiol Biotechnol. 95(3):577–91. https://doi.org/10.1007/s00253-012-4175-0
Fibriana F, Upaichit A, Cheirsilp B (2021) Turning waste into valuable products: utilization of agroindustrial oily wastes as the low-cost media for microbial lipase production. J Phys Conf Ser 1918(5):52028. https://doi.org/10.1088/1742-6596/1918/5/052028
Sandoval G, Casas-Godoy L, Bonet-Ragel K, Rodrigues J, Ferreira-Dias S, Valero F (2017) Enzyme-catalyzed production of biodiesel as alternative to chemical-catalyzed processes: advantages and constraints. Curr Biochem Eng 4(2):109–141
Toro EC, Rodríguez DF, Morales N, García LM, Godoy CA (2019) Novel combi-lipase systems for fatty acid ethyl esters production. Catalysts. https://doi.org/10.3390/catal9060546
Tongboriboon K, Cheirsilp B, H-Kittikun A (2010) Mixed lipases for efficient enzymatic synthesis of biodiesel from used palm oil and ethanol in a solvent-free system. J Mol Catal B Enzym. https://doi.org/10.1016/j.molcatb.2010.07.005
Funding
Funding for this research was provided by the Research and Development Office (Contract No. AGR5111990069S) and the Graduate School, Prince of Songkla University, Thailand, Fiscal Year 2021. The financial support for Fidia Fibriana was from the Overseas Postgraduate Scholarship (BPPLN) 2019 Program, Directorate General of Higher Education, Ministry of Education, Culture, Research and Technology of Indonesia. The second and third authors are supported by the Thailand Research Fund under Grant No. RTA6280014.
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
Author information
Authors and Affiliations
Contributions
Fidia Fibriana: Data curation, Visualization, Investigation, Software, Writing—original draft. Apichat Upaichit: Supervision, Conceptualization, Methodology, Data curation, Writing—review and editing. Benjamas Cheirsilp: Supervision, Writing—review and editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• POME can be utilized as a medium for microbial lipases production.
• A combined approach of valorization and bioremediation of POME was performed.
• The co-culture system enhanced lipases production and bioremediation performance.
• The mixed cells of yeast and bacteria improved biodiesel production.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fibriana, F., Upaichit, A. & Cheirsilp, B. Low-cost production of cell-bound lipases by pure and co-culture of yeast and bacteria in palm oil mill effluent and the applications in bioremediation and biodiesel synthesis. Biomass Conv. Bioref. 13, 10823–10844 (2023). https://doi.org/10.1007/s13399-021-02070-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-021-02070-z