Abstract
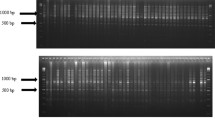
Cotton (Gossypium spp.) is the most important natural fiber and oil source worldwide. The genus Gossypium has 45 diploid and 5 allotetraploid species. Currently cotton has only 4 cultivated species, two tetraploid species [G. hirsutum L. and G. barbadense L., (2n = 4x = 52)] and two diploid species [G. arboreum L. and G. herbaceum L., (2n = 2x = 26]. Continuous artificial selection and cultivation of available cotton cultivars has led to genetic erosion and loss of useful genetic loci from the available cotton germplasm. Different molecular markers used in genetic fingerprinting of cottons, revealed narrow genetic variability. In order to provide data on genetic diversity present in our cotton germplasm collection, we carried out genetic fingerprinting study of 17 diploid and tetraploid cotton accessions by inter retrotransposon amplified polymorphism markers. The results showed low-moderate genetic variability (0.0–18%) among the studied cultivars. However, we obtained high genetic variability within each species (45–80%). Moreover, some specific bands (alleles) were identified in these species. Such moderate to high genetic variability in cotton germplasm is of high importance for choosing parental plants for hybridization. Inter retrotransposon amplified polymorphism markers could efficiently differentiate the cultivars and the studied diploid species from tetraploids. Therefore, these are suitable molecular markers for fast screening and genetic fingerprinting of large cotton germplasm.



Similar content being viewed by others
References
Abd El-Moghny AM, Max Mariz S, GibelyReham HA. Nature of genetic divergence among some cotton genotypes. J Cotton Sci. 2015;19:368–74.
Alzohairy AM, Gyulai G, Ramadan MF, Edris S, Sabir JSM, Jansen RK, Eissa H, Bahieldin A. Retrotransposon-based molecular markers for assessment of genomic diversity. Funct Plant Biol. 2014;41:781–9.
Biswas MK, Xu Q, Deng X. Utility of RAPD, ISSR, IRAP and REMAP markers for the genetic analysis of Citrus spp. Sci Hortic. 2010;124:254–61.
Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007. https://doi.org/10.1111/j.1471-8286.2007.01758.x.
Freeland JR, Kirk S, Petersen D, editors. Molecular ecology. 2nd ed. London: Wiley; 2011. p. 449.
Iqbal MJ, Aziz N, Saeed NA, Zafar Y, Malik KA. Genetic diversity evaluation of some elite cotton varieties by RAPD analysis. Theor Appl Genet. 1997;94:139–44.
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet. 1999;98:704–11.
Krizman M, Jakse J, Baricevic D, Javornik B, Mirko P. Robust CTAB-activated charcoal protocol for plant DNA extraction. Acta Agric Slov. 2006;87:427–33.
Liu D, Guo X, Lin Z, Nie Y, Zhang X. Genetic diversity of Asian cotton (Gossypium arboretum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol. 2006;53:1145–52.
Malik W, Ashraf J, Iqbal MZ, Khan AA, Qayyum A, Abid MA, Noor E, Ahmad MQ, Abbasi GH. Molecular markers and cotton genetic improvement: current status and future prospects. Cairo: Hindawi Publishing Corporation; 2014. p. 15. https://doi.org/10.1155/2014/607091 (Article ID 607091).
Meirmans PG. AMOVA-based clustering of population genetic data. J Hered. 2012;103:744–50.
Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–4.
Noormohammadi Z, Sheidai M, Foroutan M, Alishah O. Networking and Bayesian analyses of genetic affinity in cotton germplasm. Nucleus. 2015;58:33–45.
Noormohammadi Z, Shamee MH, Sheidai M. Chromosome pairing analysis of some parental and F2 cotton progenies. Nucleus. 2013;56:37–40.
Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95.
Sheidai M, Afshar F, Keshavarzi M, Talebi SM, Noormohammadi Z, Shafaf T. Genetic diversity and genome size variability in Linum austriacum (Linaceae) populations. Biochem Syst Ecol. 2014;57:20–6.
Sheidai M, Dokhanchei A, Noormohammadi Z. Karyotype and chromosome pairing analysis in some Iranian upland cotton (Gossypium hirsutum) cultivars. Cytologia. 2008;73:275–81.
Sheidai M, Shahriari ZH, Roknizadeh H, Noormohammadi Z. RAPD and cytogenetic study of some tetraploid cotton (Gossypium hirsutum L.) cultivars and their hybrids. Cytologia. 2007;72:77–82.
Sheidai M. Cytogenetic distinctiveness of sixty-six tetraploid cotton (Gossypium hirsutum L.) cultivars based on meiotic data. Acta Bot Croat. 2008;67:209–20.
Teo CH, Tan SH, Ho CL, Faridah QZ, Othman YR, Heslop-Harrison JS, Kalendar R, Schulman AH. Genome constitution and classification using retrotransposon-based markers in the orphan crop banana. J Plant Biol. 2005;48:96–105.
Tyagi P, Gore MA, Bowman DT, Campbell BT, Udall JA, Kuraparthy V. Genetic diversity and population structure in the US upland cotton (Gossypium hirsutum L.). Theor Appl Genet. 2014;127:283–95.
Ulloa M, Brubaker C, Chee P. Cotton. In: Kole C, editor. Genome mapping and molecular breeding, vol. 6. New York: Technical Crops Springer; 2007.
Van Esbroeck G, Bowman DT. Cotton germplasm diversity and its importance to cultivar development. J Cotton Sci. 1998;2:121–9.
Wang X, Ma J, Yang S, Zhang G, Ma ZY. Assessment of genetic diversity among Chinese upland cottons with Fusarium and/or Verticillium wilts resistance by AFLP and SSR markers. Front Agric China. 2007;1:129–35.
Weising K, Nybom H, Wolf K, Kahl G. DNA finger printing in plants. 2nd ed. Boca Raton: CRC Press; 2005. p. 444.
Zhang Y, Wang XF, Li ZK, Zhang GY, Ma ZY. Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. Genet Mol Resour. 2011;10:1462–70.
Acknowledgements
We acknowledge Gorgan cotton research center for providing cotton samples.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Noormohammadi, Z., Ibrahim-Khalili, N., Sheidai, M. et al. Genetic fingerprinting of diploid and tetraploid cotton cultivars by retrotransposon-based markers. Nucleus 61, 137–143 (2018). https://doi.org/10.1007/s13237-018-0237-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-018-0237-8