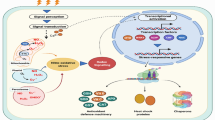
Abstract
The recent global climate change has directly impacted major biotic and abiotic stress factors affecting crop productivity worldwide. Therefore, the need of the hour is to develop sustainable multiple stress tolerant crops through modern biotechnological approaches to cope with climate change. Hybrid proline rich proteins (HyPRPs) are the cell-wall structural proteins, which contain an N-terminal repetitive proline-rich domain and a C-terminal conserved eight-cysteine motif domain. HyPRPs are known to regulate multiple abiotic and biotic stress responses in plants. Recently, a few HyPRPs have been characterized as negative regulators of abiotic and biotic stress responses in different plants. Disruption of such negative regulators for desirable positive phenotypic traits has been made possible through the advent of advanced genome engineering tools. In the past few years, CRISPR/Cas9 has emerged as a novel breakthrough technology for crop improvement by target specific editing of known negative regulatory host genes. Here, we have described the mechanism of action and the role of known HyPRPs in regulating different biotic and abiotic stress responses in major crop plants. We have also discussed the importance of the CRISPR/Cas9 based genome editing system in targeting known negative regulatory HyPRPs for multi-stress crop tolerance using the tomato crop model. Application of genome editing to manipulate the HyPRPs of major crop plants holds promise in developing newer stress management methods in this rapidly changing climate and would lead in the future to sustain crop productivity.




Similar content being viewed by others
References
Abdallah NA, Moses V, Prakash C (2014) The impact of possible climate changes on developing countries. GM Crops Food 5:77–80
Ábrahám E, Hourton-Cabassa C, Erdei L, Szabados L (2010) Methods for determination of proline in plants. In: Sunkar R (ed) Plant stress tolerance. Methods in molecular biology (methods and protocols), vol 639. Humana Press
Abudayyeh OO, Gootenberg JS, Essletzbichler P et al (2017) RNA targeting with CRISPR-Cas13. Nature 550:280–284
Aman R, Ali Z, Butt H et al (2018) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19:1
Amitai G, Sorek R (2016) CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol 14:67–76
Anders C, Niewoehner O, Duerst A, Jinek M (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513:569–573
Andersson M, Turesson H, Olsson N, Fält AS, Olsson P et al (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant 164:378–384
Bari VK, Nassar JA, Kheredin SM et al (2019) CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci Rep 9:11438
Barrangou R, Fremaux C, Deveau H et al (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Bastola DR, Pethe VV, Winicov I (1998) Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol 38:1123–1135
Belhaj K, Chaparro-Garcia A, Kamoun S et al (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Met 9:39
Bernabe-Orts JM, Casas-Rodrigo I, Minguet EG et al (2019) Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol J 17:1971–1984
Bernhardt C, Tierney ML (2000) Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol 122(3):705–714
Bin Moon S, Lee JM, Kang JG et al (2018) Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat Commun 9:3651
Bogdanove AJ, Voytas DF (2011) TAL effectors: customizable proteins for DNA targeting. Science 333:1843–1846
Boron AK, Orden JV, Markakis MN, Mouille GA, Dirk V et al (2014) Proline-rich protein-like PRPL1 controls elongation of root hairs in Arabidopsis thaliana. J Exp Bot 65:5485–5495
Breseghello F, Coelho ASG (2013) Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J Agric Food Chem 61:8277–8286
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Chikkaputtaiah C, Debbarma J, Baruah I et al (2017) Molecular genetics and functional genomics of abiotic stress-responsive genes in oilseed rape (Brassica napus L.): a review of recent advances and future. Plant Biotechnol Rep 11:365–384
Clasen BM, Stoddard TJ, Luo S, Demorest ZL et al (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 14:169–176
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B: Biol Sci 363:557–572
D’Ambrosio C, Stigliani AL, Giorio G (2018) CRISPR/Cas9 editing of carotenoid genes in tomato. Trans Res 27:367–378
Davies JP, Kumar S, Sastry-Dent L (2017) Use of zinc-finger nucleases for crop improvement program. Mol Biol Trans Sci 149:47–63
Debbarma J, Sarki YN, Saikia B et al (2019) Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol Biotechnol 61:153–172
Deltcheva E, Chylinski K, Sharma CM et al (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607
Deutch CE, Winicov I (1995) Post-transcriptional regulation of a salt-inducible alfalfa gene encoding a putative chimeric proline-rich cell wall protein. Plant Mol Biol 37017:411–418
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096
Dvorakova L, Cvckova F, Fischer L (2007) Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genom 16:1–16
Feng Z, Mao Y, Xu N et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637
Forsyth A, Weeks T, Richael C, Duan H (2016) Transcription activator-like effector nucleases (TALEN)-mediated targeted DNA insertion in potato plants. Front Plant Sci 7:1572
Fowler TJ, Bernhardt C, Tierney ML (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins 1. Plant Physiol 121:1081–1091
Fuentes A, Carlos N, Ruiz Y et al (2016) Field trial and molecular characterization of RNAi-transgenic tomato plants that exhibit resistance to tomato yellow leaf curl geminivirus. Mol Plant-Microbe Interact 29:197–209
Gaj T, Gersbach C, Barbas CF III (2013) ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trend Biotecnol 31:397–405. https://doi.org/10.1016/j.tibtech.2013.04.004.ZFN
Gangwani L, Mikrut M, Galcheva-gargova Z, Davis RJ (1998) Interaction of ZPR1 with translation elongation factor-1 in proliferating cells. J Cell Biol 143:1471–1484
Gao L, Cox DBT, Yan WX et al (2017) Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol 35:789–792
Gilchrist E, Haughn G (2010) Reverse genetics techniques: engineering loss and gain of gene function in plants. Brief Funct Genom 9:103–110
Godfray HCJ, Crute IR, Haddad L et al (2010) The future of the global food system. Philos Trans R Soc Lond Ser B Biol Sci 365:2769–2777
Goodwin W, Pallas JA, Jenkins GI (1996) Transcripts of a gene encoding a putative cell wall-plasma membrane linker protein are specifically cold-induced in Brassica napus. Plant Mol Biol 31:771–781
Gothandam KM, Flower RÁ (2010) OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol Biol 72:125–135
Gujjar RS, Karkute S, Rai A (2018) Proline-rich proteins may regulate free cellular proline levels during drought stress in tomato. Curr Sci 114:915–920
Gupta RM, Musunuru K (2014) The emergence of genome-editing technology: ZFNs, TALENs, and CRISPR-Cas9. J Clin Investig 124:4154–4161
Han Y, Teng K, Nawaz G et al (2019) Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations. 3 Biotech 9:387
He CY, Zhang JS, Chen SY (2002) A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet 104:1125–1131
He Y, Zhu M, Wang L et al (2018) Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant 11:1210–1213
Heler R, Marraffini LA, Bikard D (2014) Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol 93:1–9
Hirano S, Nishimasu H, Ishitani R, Nureki O (2016) Structural basis for the altered PAM specificities of engineered CRISPR-Cas9. Mol Cell 61:886–894
Horvath P, Romero DA, Coute-Monvoisin A-C et al (2008) Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190:1401–1412
Hsu C-T, Cheng Y-J, Yuan Y-H et al (2019) Application of Cas12a and nCas9-activation-induced cytidine deaminase for genome editing and as a non-sexual strategy to generate homozygous/multiplex edited plants in the allotetraploid genome of tobacco. Plant Mol Biol 101:355–371
Hu N, Xian Z, Li N et al (2019) Rapid and user-friendly open-source CRISPR/Cas9 system for single- or multi-site editing of tomato genome. Hort Res 6:1–14
Huang G, Gong S, Xu W, Li P et al (2011) GhHyPRP4, a cotton gene encoding putative hybrid proline-rich protein is preferentially expressed in leaves and involved in plant response to cold stress. Acta Biochim Biophys Sin 43:519–527
Javier M, Susana U, Giusto NM et al (2016) Saline and osmotic stresses stimulate PLD/diacylglycerol kinase activities and increase the level of phosphatidic acid and proline in barley roots. Environ Exp Bot. 128:69–78
Jinek M, Jiang F, Taylor DW et al (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997
Jose M (2005) Expression of the promoter of HyPRP, an embryo-specific gene from Zea mays in maize and tobacco transgenic plants. Gene 356:146–152
Josè-estanyol M, Puigdomènech P (2000) Plant cell wall glycoproteins and their genes. Plant Physiol Biochem 38:97–108
Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT (2009) Priming in systemic plant immunity. Science 324(5923):89–91
Khan AO, White CW, Pike JA et al (2019) Optimised insert design for improved single-molecule imaging and quantification through CRISPR-Cas9 mediated knock-in. Sci Rep 9:14219
Kim H, Kim ST, Ryu J, Kang BC et al (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:14406
Kim D, Alptekin B, Budak H (2018) CRISPR/Cas9 genome editing in wheat. Funct Integr Genom 18:31–41
Kleinstiver BP, Tsai SQ, Prew MS et al (2016) Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34:869–874
Krumbach K, Sonntag CK, Eggeling L, Marienhagen J (2019) CRISPR/Cas12a mediated genome editing to introduce amino acid substitutions into the mechanosensitive channel MscCG of Corynebacterium glutamicum. ACS Synth Biol 8:2726–2734
Lamaoui M, Jemo M, Datla R, Bekkaoui F (2018) Heat and drought stresses in crops and approaches for their mitigation. Front Chem 6:1–14
Lee JK, Jeong E, Lee J et al (2018) Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 9:3048
Li J, Ouyang B, Wang T et al (2016) HyPRP1 gene suppressed by multiple stresses plays a negative role in abiotic stress tolerance in tomato. Front Plant Sci 7:1–14
Li R, Liu C, Zhao R et al (2019) CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol 19:38
Liang Z, Chen K, Li T et al (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261
Liang M, Li Z, Wang W et al (2019) A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat Commun 10:3672
Lindbo JA (2012) A historical overview of RNAi in plants. Methods Mol Biol 894:1–16
Liu D, Chen X, Liu J, Ye J, Guo Z (2012) The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot 63:3899–3912
Liu H, Sultan MARF, Liu XL et al (2015) Physiological and comparative proteomic analysis reveals different drought responses in roots and leaves of drought-tolerant wild wheat (Triticum boeoticum). PLoS ONE 10:1–29
Liu H, Ding Y, Zhou Y et al (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10:530–532
Makarova KS, Wolf YI, Alkhnbashi OS et al (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736
Manghwar H, Lindsey K, Zhang X, Jin S (2019) CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci 24:1102–1125
Marraffini LA (2015) CRISPR-Cas immunity in prokaryotes. Nature 526:55–61
Marwein R, Debbarma J, Sarki YN, Baruah I et al (2019) Genetic engineering/genome editing approaches to modulate signaling processes in abiotic stress tolerance. In: Iqbal M (ed) Plant signaling molecules. Elsevier, Amsterdam, pp 63–82
Mellacheruvu S, Tamirisa S, Vudem DR, Khareedu VR (2016) Pigeonpea hybrid-proline-rich protein (CcHyPRP) confers biotic and abiotic stress tolerance in transgenic rice. Front Plant Sci 6:1–13
Mojica FJM, Rodriguez-Valera F (2016) The discovery of CRISPR in archaea and bacteria. FEBS J 283:3162–3169
Musunuru K (2017) Genome editing: the recent history and perspective in cardiovascular diseases. J Am Coll Cardiol 70:2808–2821
Nadakuduti SS, Buell CR, Voytas D, Starker C et al (2018) Genome editing for crop improvement—applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front Plant Sci 9:1607
Neto LB, De Oliveira RR, Wiebke-strohm B et al (2013) Identification of the soybean HyPRP family and specific gene response to Asian soybean rust disease. Genet Mol Biol 224:214–224
Otte O, Barz W (2000) Characterization and oxidative in vitro cross-linking of an extensin-like protein and a proline-rich protein purified from chickpea cell walls. Phytochemistry 53:1–5
Pan C, Ye L, Qin L et al (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:2–10
Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M (2017) Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci 8:537
Peer R, Rivlin G, Golobovitch S et al (2015) Targeted mutagenesis using zinc-finger nucleases in perennial fruit trees. Planta 241:941–951
Peng T, Jia M-M, Liu J-H (2015) RNAi-based functional elucidation of PtrPRP, a gene encoding a hybrid proline rich protein, in cold tolerance of Poncirus trifoliata. Front Plant Sci 6:1–12
Peters K, Breitsameter L, Gerowitt B (2014) Impact of climate change on weeds in agriculture: a review. Agron Sustain Dev 34:707–721
Priyanka B, Sekhar K, Reddy VD, Rao KV (2010) Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol J 8:76–87
Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36:1–8
Ramu VS, Swetha TN, Sheela SH et al (2016) Simultaneous expression of regulatory genes associated with specific drought-adaptive traits improves drought adaptation in peanut. Plant Biotechnol J 14:1008–1020
Ran Y, Patron N, Kay P et al (2018) Zinc finger nuclease-mediated precision genome editing of an endogenous gene in hexaploid bread wheat (Triticum aestivum) using a DNA repair template. Plant Biotechnol J 16:2088–2101
Ruchita S, Rohit S (2017) Effect of global warming on Indian agriculture. J Climatol Weather Forecast 5:1–5
Sanchez-Leon S, Gil-Humanes J, Ozuna CV et al (2018) Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J 16:902–910
Schiml S, Fauser F, Puchta H (2016) CRISPR/Cas-mediated site-specific mutagenesis in Arabidopsis thaliana using Cas9 nucleases and paired nickases. Methods Mol Biol 1469:111–122
Selma S, Bernabe-Orts JM, Vazquez-Vilar M et al (2019) Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol J 17:1703–1705
Shan Q, Wang Y, Li J et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotecnol 31:686–688
Shan Q, Baltes NJ, Atkins P et al (2018) ZFN, TALEN and CRISPR-Cas9 mediated homology directed gene insertion in Arabidopsis: a disconnect between somatic and germinal cells. J Genet Genom 45:681–684
Shmakov S, Abudayyeh OO, Makarova KS et al (2015) Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60:385–397
Singh D, Chaudhary S, Kumar R et al (2016) RNA interference technology—applications and limitations. InTech, Croatia
Soyars CL, Peterson BA, Burr CA, Nimchuk ZL (2018) Cutting edge genetics: CRISPR/Cas9 editing of plant genomes. Plant Cell Physiol 59:1608–1620
Sundaresan S, Philosoph-hadas S, Ma C et al (2018) The tomato hybrid proline-rich protein regulates the abscission zone competence to respond to ethylene signals. Hortic Res 5:28
Tan J, Zhuo C, Guo Z (2013) Nitric oxide mediates cold- and dehydration-induced expression of a novel MfHyPRP that confers tolerance to abiotic stress. Physiol Plant 149(3):310–320
Tang X, Lowder LG, Zhang T et al (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17103
Tian S, Jiang L, Gao Q et al (2017) Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep 36:399–406
Veillet F, Perrot L, Chauvin L et al (2019) Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci 20:402
Weyman PD, Pan Z, Feng Q, Gilchrist DG, Bostock RM (2006) DEA 1, a circadian- and cold-regulated tomato gene, protects yeast cells from freezing death. Plant Mol Biol 62(4–5):547–559
Winicov I, Bastola DR (1999) Transgenic overexpression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants 1. Plant Physiol 120:473–480
Wolter F, Puchta H (2018) The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J Cell Mol Biol 94:767–775
Wu X, Scott DA, Kriz AJ et al (2014) Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32:670–676
Xiao G, Zhang S, Liang Z et al (2019) Identification of Mycobacterium abscessus species and subspecies using the Cas12a/sgRNA-based nucleic acid detection platform. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-019-03757-y
Yan WX, Mirzazadeh R, Garnerone S et al (2017) BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8:15058
Yang J, Zhang Y, Wang X et al (2018) HyPRP1 performs a role in negatively regulating cotton resistance to V. dahliae via the thickening of cell walls and ROS accumulation. BMC Plant 1–18:339
Yeom S, Seo E, Oh S et al (2012) A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J 69:755–768
Zaidi SS-E-A, Mahfouz MM, Mansoor S (2017) CRISPR-Cpf1: a new tool for plant genome editing. Trends Plant Sci 22:550–553
Zegaoui Z, Boumediene TH, Planchais S (2017) Variation in relative water content, proline accumulation and stress gene expression in two cowpea landraces under drought. J Plant Physiol 218:26–34
Zhang Y, Liang Z, Zong Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617
Zhang S, Zhang R, Gao J et al (2019) Highly efficient and heritable targeted mutagenesis in wheat via the Agrobacterium tumefaciens-mediated CRISPR/Cas9 system. Int J Mol Sci 20:4257
Zhou J, Xin X, He Y et al (2019) Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep 38:475–485
Acknowledgements
Authors would like to acknowledge Science and Engineering Research Board (SERB), Govt. of India for the financial support to C. C. in the form of Ramanujan Fellowship (SB/S2/RJN-078/2014) and Early Career Research Award (ECR/2016/001288).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors ensure that the presented work followed accepted principles of ethical and professional conduct. We hereby confirm that the authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Saikia, B., Singh, S., Debbarma, J. et al. Multigene CRISPR/Cas9 genome editing of hybrid proline rich proteins (HyPRPs) for sustainable multi-stress tolerance in crops: the review of a promising approach. Physiol Mol Biol Plants 26, 857–869 (2020). https://doi.org/10.1007/s12298-020-00782-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00782-6