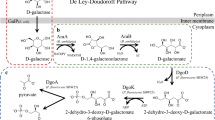
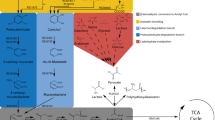
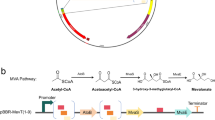
Abstract
Lignin, a complex aromatic polymer, is a structural component of plant biomass. It decomposes with difficulty because of its rigidity properties, however, lignin valorization is essential for the economics of lignocellulosic biorefineries. Pseudomonas putida has been extensively investigated as a promising host strain for lignin valorizations due to intrinsic traits, such as low nutritional requirement, high tolerance to toxicity, and metabolic versatility with a wide spectrum of substrates, such as aromatic compounds. Although a naturally occurring, lignin-utilizing P. putida strain has been reported, it is necessary to engineer the genome of P. putida for efficient lignin utilization. For biological lignin valorization, the decomposition of lignin polymer to low-molecular weight compounds and transformation of lignin-derived aromatic compounds to value-added chemicals is essential. Various tools of synthetic biology have been developed for the genome engineering of P. putida; efforts in metabolic engineering have been made to expand aromatic substrate specificity and to improve productivity of value-added chemicals. Development of high-performance bio-parts and biosensors for lignin valorization has also been done. In this review, we present recent research on genome engineering tools developed for P. putida and metabolic engineering employed in P. putida to improve lignin valorization.
Similar content being viewed by others
References
Nikel, P. I., E. Martínez-García, and V. de Lorenzo (2014) Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 12: 368–379.
Vanholme, R., B. Demedts, K. Morreel, J. Ralph, and W. Boerjan (2010) Lignin biosynthesis and structure. Plant Physiol. 153: 895–905.
Ezeji, T., N. Qureshi, and H. P. Blaschek (2007) Butanol production from agricultural residues: impact of degradation products on Clostridium beijerinckii growth and butanol fermentation. Biotechnol. Bioeng. 97: 1460–1469.
Bugg, T. D. H. and R. Rahmanpour (2015) Enzymatic conversion of lignin into renewable chemicals. Curr. Opin. Chem. Biol. 29: 10–17.
Lee, S., M. Kang, J. H. Bae, J. H. Sohn, and B. H. Sung (2019) Bacterial valorization of lignin: strains, enzymes, conversion pathways, biosensors, and perspectives. Front. Bioeng. Biotechnol. 7: 209.
Santos, A., S. Mendes, V. Brissos, and L. O. Martins (2014) New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl. Microbiol. Biotechnol. 98: 2053–2065.
Jiménez, J. I., B. Miñambres, J. L. García, and E. Díaz (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4: 824–841.
Min, K., G. Gong, H. M. Woo, Y. Kim, and Y. Um (2015) A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Sci. Rep. 5: 8245.
Salvachúa, D., E. M. Karp, C. T. Nimlos, D. R. Vardon, and G. T. Beckham (2015) Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 17: 4951–4967.
Linger, J. G., D. R. Vardon, M. T. Guarnieri, E. M. Karp, G. B. Hunsinger, M. A. Franden, C. W. Johnson, G. Chupka, T. J. Strathmann, P. T. Pienkos, and G. T. Beckham (2014) Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. USA. 111: 12013–12018.
Lin, L., Y. Cheng, Y. Pu, S. Sun, X. Li, M. Jin, E. A. Pierson, D. C. Gross, B. E. Dale, S. Y. Dai, A. J. Ragauskas, and J. S. Yuan (2016) Systems biology-guided biodesign of consolidated lignin conversion. Green Chem. 18: 5536–5547.
Nikel, P. I. and V. de Lorenzo (2018) Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab. Eng. 50: 142–155.
Domröse, A., A. S. Klein, J. Hage-Hülsmann, S. Thies, V. Svensson, T. Classen, J. Pietruszka, K. E. Jaeger, T. Drepper, and A. Loeschcke (2015) Efficient recombinant production of prodigiosin in Pseudomonas putida. Front. Microbiol. 6: 972.
Martínez-García, E. and V. de Lorenzo (2011) Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13: 2702–2716.
Chen, Z., W. Ling, and G. Shang (2016) Recombineering and I-SceI-mediated Pseudomonas putida KT2440 scarless gene deletion. FEMS Microbiol. Lett. 363: fnw231.
Martínez-García, E., P. I. Nikel, M. Chavarría, and V. de Lorenzo (2014) The metabolic cost of flagellar motion in Pseudomonas putida KT 2440. Environ. Microbiol. 16: 291–303.
Salvachúa, D., T. Rydzak, R. Auwae, A. De Capite, B. A. Black, J. T. Bouvier, N. S. Cleveland, J. R. Elmore, J. D. Huenemann, R. Katahira, W. E. Michener, D. J. Peterson, H. Rohrer, D. R. Vardon, G. T. Beckham, and A. M. Guss (2020) Metabolic engineering of Pseudomonas putida for increased polyhydroxyalkanoate production from lignin. Microb. Biotechnol. 13: 290–298.
Luo, X., Y. Yang, W. Ling, H. Zhuang, Q. Li, and G. Shang (2016) Pseudomonas putida KT2440 markerless gene deletion using a combination of λ Red recombineering and Cre/loxP site-specific recombination. FEMS Microbiol. Lett. 363: fnw014.
Choi, K. R. and S. Y. Lee (2020) Protocols for RecET-based markerless gene knockout and integration to express heterologous biosynthetic gene clusters in Pseudomonas putida. Microb. Biotechnol. 13: 199–209.
Choi, K. R., J. S. Cho, I. J. Cho, D. Park, and S. Y. Lee (2018) Markerless gene knockout and integration to express heterologous biosynthetic gene clusters in Pseudomonas putida. Metab. Eng. 47: 463–474.
Martínez-García, E. and V. de Lorenzo (2017) Molecular tools and emerging strategies for deep genetic/genomic refactoring of Pseudomonas. Curr. Opin. Biotechnol. 47: 120–132.
Sun, J., Q. Wang, Y. Jiang, Z. Wen, L. Yang, J. Wu, and S. Yang (2018) Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb. Cell Fact. 17: 41.
Tan, S. Z., C. R. Reisch, and K. L. Prather (2018) A robust CRISPR interference gene repression system in Pseudomonas. J. Bacteriol. 200: e00575–00517.
Mougiakos, I., P. Mohanraju, E. F. Bosma, V. Vrouwe, M. F. Bou, M. I. Naduthodi, A. Gussak, R. B. Brinkman, R. Van Kranenburg, and J. Van Der Oost (2017) Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat. Commun. 8: 1647.
Aparicio, T., V. de Lorenzo, and E. Martínez-García (2018) CRISPR/Cas9-based counterselection boosts recombineering efficiency in Pseudomonas putida. Biotechnol. J. 13: e1700161.
Lieder, S., P. I. Nikel, V. de Lorenzo, and R. Takors (2015) Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microb. Cell Fact. 14: 23.
Martínez-García, E., P. I. Nikel, T. Aparicio, and V. de Lorenzo (2014) Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Fact. 13: 159.
Wang, J., W. Ma, Y. Wang, L. Lin, T. Wang, Y. Wang, Y. Li, and X. Wang (2018) Deletion of 76 genes relevant to flagella and pili formation to facilitate polyhydroxyalkanoate production in Pseudomonas putida. Appl. Microbiol. Biotechnol. 102: 10523–10539.
García-Hidalgo, J., K. Ravi, L. L. Kuré, G. Lidén, and M. Gorwa-Grauslund (2019) Identification of the two-component guaiacol demethylase system from Rhodococcus rhodochrous and expression in Pseudomonas putida EM42 for guaiacol assimilation. AMB Express. 9: 34.
de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis (1993) Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 123: 17–24.
Pérez-Martin, J. and V. de Lorenzo (1996) VTR expression cassettes for engineering conditional phenotypes in Pseudomonas: activity of the Pu promoter of the TOL plasmid under limiting concentrations of the XylR activator protein. Gene. 172: 81–86.
de Lorenzo, V., S. Fernández, M. Herrero, U. Jakubzik, and K. N. Timmis (1993) Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene. 130: 41–46.
Gawin, A., S. Valla, and T. Brautaset (2017) The XylS/Pm regulator/promoter system and its use in fundamental studies of bacterial gene expression, recombinant protein production and metabolic engineering. Microb. Biotechnol. 10: 702–718.
Wittgens, A., B. Santiago-Schuebel, M. Henkel, T. Tiso, L. M. Blank, R. Hausmann, D. Hofmann, S. Wilhelm, K. E. Jaeger, and F. Rosenau (2018) Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida-a step forward to tailor-made rhamnolipids. Appl. Microbiol. Biotechnol. 102: 1229–1239.
Silva-Rocha, R., E. Martínez-García, B. Calles, M. Chavarría, A. Arce-Rodríguez, A. de Las Heras, A. D. Paez-Espino, G. Durante-Rodríguez, J. Kim, P. I. Nikel, R. Platero, and V. de Lorenzo (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41: D666–D675.
Martínez-García, E., T. Aparicio, A. Goñi-Moreno, S. Fraile, and V. de Lorenzo (2015) SEVA 2.0: an update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 43: D1183–D1189.
Martínez-García, E., A. Goñi-Moreno, B. Bartley, J. McLaughlin, L. Sánchez-Sampedro, H. Pascual del Pozo, C. P. Hernández, A. S. Marletta, D. De Lucrezia, G. Sánchez-Fernández, S. Fraile, and V. de Lorenzo (2020) SEVA 3.0: an update of the Standard European Vector Architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic Acids Res. 48: D1164–D1170.
Bugg, T. D. H., J. J. Williamson, and G. M. M. Rashid (2020) Bacterial enzymes for lignin depolymerisation: new biocatalysts for generation of renewable chemicals from biomass. Curr. Opin. Chem. Biol. 55: 26–33.
Choudhary, A., H. Purohit, and P. S. Phale (2017) Benzoate transport in Pseudomonas putida CSV86. FEMS Microbiol. Lett. 364: fnx118.
Nichols, N. N. and C. S. Harwood (1997) PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179: 5056–5061.
Tan, K., C. Chang, M. Cuff, J. Osipiuk, E. Landorf, J. C. Mack, S. Zerbs, A. Joachimiak, and F. R. Collart (2013) Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids. Proteins. 81: 1709–1726.
Fujita, M., K. Mori, H. Hara, S. Hishiyama, N. Kamimura, and E. Masai (2019) A TonB-dependent receptor constitutes the outer membrane transport system for a lignin-derived aromatic compound. Commun. Biol. 2: 432.
Vermaas, J. V., R. A. Dixon, F. Chen, S. D. Mansfield, W. Boerjan, J. Ralph, M. F. Crowley, and G. T. Beckham (2019) Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. USA. 116: 23117–23123.
Beckham, G. T., C. W. Johnson, E. M. Karp, D. Salvachúa, and D. R. Vardon (2016) Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 42: 40–53.
Singh, R., P. Singh, and R. Sharma (2014) Microorganism as a tool of bioremediation technology for cleaning environment: a review. Proc. Int. Acad. Ecol. Environ. Sci. 4: 1–6.
Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martínez, and F. Rojo (2004) The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186: 1337–1344.
Hernández-Arranz, S., R. Moreno, and F. Rojo (2013) The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi-tier regulation strategy. Environ. Microbiol. 15: 227–241.
Johnson, C. W., P. E. Abraham, J. G. Linger, P. Khanna, R. L. Hettich, and G. T. Beckham (2017) Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab. Eng. Commun. 5: 19–25.
Abdelaziz, O. Y., K. Li, P. Tunå, and C. P. Hulteberg (2018) Continuous catalytic depolymerisation and conversion of industrial kraft lignin into low-molecular-weight aromatics. Biomass Conv. Bioref. 8: 455–470.
Mosier, N., C. Wyman, B. Dale, R. Elander, Y. Y. Lee, M. Holtzapple, and M. Ladisch (2005) Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour. Technol. 96: 673–686.
Banerjee, S., G. Mishra, and A. Roy (2019) Metabolic engineering of bacteria for renewable bioethanol production from cellulosic biomass. Biotechnol. Bioprocess Eng. 24: 713–733.
Puchałka, J., M. A. Oberhardt, M. Godinho, A. Bielecka, D. Regenhardt, K. N. Timmis, J. A. Papin, and V. A. P. M. Dos Santos (2008) Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput. Biol. 4: e1000210.
Basu, A., S. K. Apte, and P. S. Phale (2006) Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl. Environ. Microbiol. 72: 2226–2230.
Dvořák, P. and V. de Lorenzo (2018) Refactoring the upper sugar metabolism of Pseudomonas putida for co-utilization of cellobiose, xylose, and glucose. Metab. Eng. 48: 94–108.
Vinuselvi, P. and S. K. Lee (2012) Engineered Escherichia coli capable of co-utilization of cellobiose and xylose. Enzyme Microb. Technol. 50: 1–4.
Lee, J., J. N. Saddler, Y. Um, and H. M. Woo (2016) Adaptive evolution and metabolic engineering of a cellobiose- and xylose-negative Corynebacterium glutamicum that co-utilizes cellobiose and xylose. Microb. Cell Fact. 15: 20.
Wang, Y., F. Horlamus, M. Henkel, F. Kovacic, S. Schläfle, R. Hausmann, A. Wittgens, and F. Rosenau (2019) Growth of engineered Pseudomonas putida KT2440 on glucose, xylose, and arabinose: Hemicellulose hydrolysates and their major sugars as sustainable carbon sources. GCB Bioenergy. 11: 249–259.
Sánchez-Pascuala, A., L. Fernandez-Cabezon, V. de Lorenzo, and P. I. Nikel (2019) Functional implementation of a linear glycolysis for sugar catabolism in Pseudomonas putida. Metab. Eng. 54: 200–211.
Averesch, N. J. H. and J. O. Krömer (2018) Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds-present and future strain construction strategies. Front. Bioeng. Biotechnol. 6: 32.
Johnson, C. W., D. Salvachúa, P. Khanna, H. Smith, D. J. Peterson, and G. T. Beckham (2016) Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Met. Eng. Commun. 3: 111–119.
Jung, H. M., M. Y. Jung, and M. K. Oh (2015) Metabolic engineering of Klebsiella pneumoniae for the production of cis,cis-muconic acid. Appl. Microbiol. Biotechnol. 99: 5217–5225.
Lee, J. H. and V. F. Wendisch (2017) Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 257: 211–221.
Lee, S., D. Nam, J. Y. Jung, M. K. Oh, B. I. Sang, and R. J. Mitchell (2012) Identification of Escherichia coli biomarkers responsive to various lignin-hydrolysate compounds. Bioresour. Technol. 114: 450–456.
da Silva, E. A. B., M. Zabkova, J. D. Araújo, C. A. Cateto, M. F. Barreiro, M. N. Belgacem, and A. E. Rodrigues (2009) An integrated process to produce vanillin and lignin-based polyurethanes from Kraft lignin. Chem. Eng. Res. Des. 87: 1276–1292.
Sainsbury, P. D., E. M. Hardiman, M. Ahmad, H. Otani, N. Seghezzi, L. D. Eltis, and T. D. H. Bugg (2013) Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. ACS Chem. Biol. 8: 2151–2156.
Graf, N. and J. Altenbuchner (2014) Genetic engineering of Pseudomonas putida KT2440 for rapid and high-yield production of vanillin from ferulic acid. Appl. Microbiol. Biotechnol. 98: 137–149.
Rodriguez, A., D. Salvachúa, R. Katahira, B. A. Black, N. S. Cleveland, M. Reed, H. Smith, E. E. K. Baidoo, J. D. Keasling, B. A. Simmons, G. T. Beckham, and J. M. Gladden (2017) Base-catalyzed depolymerization of solid lignin-rich streams enables microbial conversion. ACS Sustainable Chem. Eng. 5: 8171–8180.
Van Duuren, J. B. J. H., D. Wijte, A. Leprince, B. Karge, J. Puchałka, J. Wery, V. A. P. M. Dos Santos, G. Eggink, and A. E. Mars (2011) Generation of a catR deficient mutant of P. putida KT2440 that produces cis, cis-muconate from benzoate at high rate and yield. J. Biotechnol. 156: 163–172.
Vardon, D. R., M. A. Franden, C. W. Johnson, E. M. Karp, M. T. Guarnieri, J. G. Linger, M. J. Salm, T. J. Strathmann, and G. T. Beckham (2015) Adipic acid production from lignin. Energy Envrion. Sci. 8: 617–628.
Salvachúa, D., C. W. Johnson, C. A. Singer, H. Rohrer, D. J. Peterson, B. A. Black, A. Knapp, and G. T. Beckham (2018) Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem. 20: 5007–5019.
Brodin, M., M. Vallejos, M. T. Opedal, M. C. Area, and G. Chinga-Carrasco (2017) Lignocellulosics as sustainable resources for production of bioplastics — A review. J. Clean. Prod. 162: 646–664.
Zhang, X., R. Luo, Z. Wang, Y. Deng, and G. Q. Chen (2009) Application of (R)-3-hydroxyalkanoate methyl esters derived from microbial polyhydroxyalkanoates as novel biofuels. Biomacromolecules. 10: 707–711.
Wang, S. Y., Z. Wang, M. M. Liu, Y. Xu, X. J. Zhang, and G. Q. Chen (2010) Properties of a new gasoline oxygenate blend component: 3-hydroxybutyrate methyl ester produced from bacterial poly-3-hydroxybutyrate. Biomass Bioenergy. 34: 1216–1222.
Tomizawa, S., J. A. Chuah, K. Matsumoto, Y. Doi, and K. Numata (2014) Understanding the limitations in the biosynthesis of polyhydroxyalkanoate (PHA) from lignin derivatives. ACS Sustainable Chem. Eng. 2: 1106–1113.
Wang, X., L. Lin, J. Dong, J. Ling, W. Wang, H. Wang, Z. Zhang, and X. Yu (2018) Simultaneous improvements of Pseudomonas cell growth and polyhydroxyalkanoate production from a lignin derivative for lignin-consolidated bioprocessing. Appl. Environ. Microbiol. 84: e01469–01418.
Wang, H. H., X. R. Zhou, Q. Liu, and G. Q. Chen (2011) Biosynthesis of polyhydroxyalkanoate homopolymers by Pseudomonas putida. Appl. Microbiol. Biotechnol. 89: 1497–1507.
Tran, T. T. and T. C. Charles (2020) Lactic acid containing polymers produced in engineered Sinorhizobium meliloti and Pseudomonas putida. PLoS One. 15: e0218302.
Eş, I., A. M. Khaneghah, F. J. Barba, J. A. Saraiva, A. S. Sant’Ana, and S. M. B. Hashemi (2018) Recent advancements in lactic acid production-a review. Food Res. Int. 107: 763–770.
Johnson, C. W. and G. T. Beckham (2015) Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Met. Eng. 28: 240–247.
Yang, J., J. H. Son, H. Kim, S. Cho, J. Na, Y. J. Yeon, and J. Lee (2019) Mevalonate production from ethanol by direct conversion through acetyl-CoA using recombinant Pseudomonas putida, a novel biocatalyst for terpenoid production. Microb. Cell Fact. 18: 168.
Lee, J. H., S. Lama, J. R. Kim, and S. H. Park (2018) Production of 1,3-propanediol from glucose by recombinant Escherichia coli BL21 (DE3). Biotechnol. Bioprocess Eng. 23: 250–258.
Nijkamp, K., R. G. M. Westerhof, H. Ballerstedt, J. A. M. De Bont, and J. Wery (2007) Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p-coumarate from glucose. Appl. Microbiol. Biotechnol. 74: 617–624.
Yu, S., M. R. Plan, G. Winter, and J. O. Krömer (2016) Metabolic engineering of Pseudomonas putida KT2440 for the production of para-hydroxy benzoic acid. Front. Bioeng. Biotechnol. 4: 90.
Wierckx, N. J. P., H. Ballerstedt, J. A. M. de Bont, and J. Wery (2005) Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71: 8221–8227.
Beller, H. R., A. V. Rodrigues, K. Zargar, Y. W. Wu, A. K. Saini, R. M. Saville, J. H. Pereira, P. D. Adams, S. G. Tringe, C. J. Petzold, and J. D. Keasling (2018) Discovery of enzymes for toluene synthesis from anoxic microbial communities. Nat. Chem. Biol. 14: 451–457.
Kwon, K. K., D. H. Lee, S. J. Kim, S. L. Choi, E. Rha, S. J. Yeom, B. Subhadra, J. Lee, K. J. Jeong, and S. G. Lee (2018) Evolution of enzymes with new specificity by high-throughput screening using DmpR-based genetic circuits and multiple flow cytometry rounds. Sci. Rep. 8: 2659.
Ho, J. C. H., S. V. Pawar, S. J. Hallam, and V. G. Yadav (2018) An improved whole-cell biosensor for the discovery of lignintransforming enzymes in functional metagenomic screens. ACS Synth. Biol. 7: 392–398.
Strachan, C. R., R. Singh, D. VanInsberghe, K. Ievdokymenko, K. Budwill, W. W. Mohn, L. D. Eltis, and S. J. Hallam (2014) Metagenomic scaffolds enable combinatorial lignin transformation. Proc. Natl. Acad. Sci. USA. 111: 10143–10148.
Jha, R. K., J. M. Bingen, C. W. Johnson, T. L. Kern, P. Khanna, D. S. Trettel, C. E. M. Strauss, G. T. Beckham, and T. Dale (2018) A protocatechuate biosensor for Pseudomonas putida KT2440 via promoter and protein evolution. Metab. Eng. Commun. 6: 33–38.
Kim, H. J., H. Jeong, and S. J. Lee (2018) Synthetic biology for microbial heavy metal biosensors. Anal. Bioanal. Chem. 410: 1191–1203.
Nguyen, N. H., J. R. Kim, and S. Park (2018) Application of transcription factor-based 3-hydroxypropionic acid biosensor. Biotechnol. Bioprocess Eng. 23: 564–572.
Gui, Q., T. Lawson, S. Shan, L. Yan, and Y. Liu (2017) The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors. 17: 1623.
Kohlstedt, M., S. Starck, N. Barton, J. Stolzenberger, M. Selzer, K. Mehlmann, R. Schneider, D. Pleissner, J. Rinkel, J. S. Dickschat, J. Venus, J. B. J. H. van Duuren, and C. Wittmann (2018) From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 47: 279–293.
Acknowledgements
This work was supported by the Intelligent Synthetic Biology Center of the Global Frontier Program (2015M3A6A8065831), the Bio & Medical Technology Development Program (2018M3A9H3024746), and the Basic Science Research Program (2019R1A2C1090726) through the National Research Foundation, and the Research Initiative Program of KRIBB.
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lee, S., Sohn, JH., Bae, JH. et al. Current Status of Pseudomonas putida Engineering for Lignin Valorization. Biotechnol Bioproc E 25, 862–871 (2020). https://doi.org/10.1007/s12257-020-0029-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0029-2