Abstract
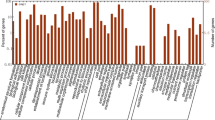
As a native in Southeast Asia, Sagittaria trifolia L. var. sinensis (arrowhead) is an aquatic perennial herb of Alismataceae family. For the aim of discovering genes and development molecular markers, high-throughput transcriptome sequencing is applied to generate enormous transcript sequences from arrowhead leaf generate. Through trimmed and assembled, 51, 836 contigs were obtained with mean length of 680 bp. Sequence similarity analyses against four public databases (NR, GO, KEGG, KOG) found 23, 595 contigs that could be annotated with gene descriptions, conserved protein domains, or gene ontology terms. In this study, 3, 861 simple sequence repeat (SSR) markers of arrowhead were identified in 3, 174 contigs of transcriptome data,and 2, 476 primer pairs were designed for marker development. Of these, 100 primers were randomly selected, synthesized and used for validation of the amplification and assessment of polymorphisms in 79 arrowhead accessions. 78 loci reliably amplified a clear single band of expexted size, and 68 loci were scoreable and polymorphic with 2 to 11 alleles per locus. The polymorphism information content valued ranged from 0.01 to 0.84. The SSR markers developed from arrowhead, could be successfully applied to Sagittaria pygmaea, with a transferability rate of 91.14% (72/79). The phylogenetic tree was also constructed to analyse the genetic diversity in arrowhead accessions. The large number of SSR markers developed in this study will be useful in the researches of population genetics, genetic diversity, and germplasm characterization in arrowhead.




Similar content being viewed by others
References
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theoretical and Applied Genetics 114:359
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32:314
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156:847–854
Chen J (1989) Systematic and evolutionary biology studies on Chinese Sagittaria. Wuhan University Press, Wuhan University Press
Chen Z, Xue C, Zhu S, Zhou F, Xuefeng LB, Liu G, Chen L (2005) GoPipe: streamlined gene ontology annotation for batch anonymous sequences with statistics Sheng wu hua xue yu sheng wu wu li jin zhan 32:187–191
Chen J-M, Gituru WR, Wang Q-F (2007) A comparison of the extent of genetic variation in the endangered Sagittaria natans and its widespread congener S. trifolia. Aquatic Botany 87:1–6
Chen J-M, Liu F, Wang Q-F, Motley TJ (2008) Phylogeography of a marsh herb Sagittaria trifolia (Alismataceae) in China inferred from cpDNA atpB–rbcL intergenic spacers. Molecular phylogenetics and evolution 48:168–175
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Du G, Yi Q, Chen J (1998) Phylogenetic relationship of Chinese Sagittaria species (Alismataceae) based on AP-PCR analysis. Acta Phytotaxonomica Sinica 36:216–221
Dutta S et al (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol 11:17
Ellis J, Burke J (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125
Eujayl I, Sorrells M, Baum M, Wolters P, Powell W (2001) Assessment of genotypic variation among cultivated durum wheat based on EST-SSRs and genomic SSRs. Euphytica 119:39–43
Feng S, Li W, Huang H, Wang J, Wu Y (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23:85–97
Grabherr MG et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology 29:644
Graham J, Smith K, MacKenzie K, Jorgenson L, Hackett C, Powell W (2004) The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theoretical and Applied Genetics 109:740–749
Han YC, Teng CZ, Hu ZL, Song YC (2008) An optimal method of DNA silver staining in polyacrylamide gels. Electrophoresis 29:1355–1358
Jiang L, Wang L, Liu L, Zhu X, Zhai L, Gong Y (2012) Development and characterization of cDNA library based novel EST-SSR marker in radish (Raphanus sativus L). Scientia Horticulturae 140:164–172
Jung S, Abbott A, Jesudurai C, Tomkins J, Main D (2005) Frequency, type, distribution and annotation of simple sequence repeats in Rosaceae ESTs. Functional & Integrative Genomics 5:136–143
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Molecular Biology 48:501–510
Kosman E, Leonard K (2005) Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol Ecol 14:415–424
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998
Liang X, Chen X, Hong Y, Liu H, Zhou G, Li S, Guo B (2009) Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biology 9:35
Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol 43:718–725
Pavlíek A (1999) Free tree-freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Prague) 45:97–99
Peng J, Lapitan NL (2005) Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Functional & Integrative Genomics 5:80–96
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15:8–15
Preiss J (1991) Biology and molecular biology of starch synthesis and its regulation Oxford surveys of plant molecular and cell biology
Rohlf FJ (1988) NTSYS-pc: numerical taxonomy and multivariate analysis system. Exeter Publishing,
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Bioinformatics methods and protocols. Springer, pp 365–386
Tan B, Liu K, Yue X-L, Liu F, Chen J-M, Wang Q-F (2008) Chloroplast DNA variation and phylogeographic patterns in the Chinese endemic marsh herb Sagittaria potamogetifolia. Aquatic Botany 89:372–378
Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L). Theoretical and Applied Genetics 106:411–422
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. TRENDS in Biotechnology 23:48–55
Wang Z et al (2010) De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genomics 11:726
Wei W et al (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 12:451
Wu ZH, Wang SZ, Hu JH, Li F, Ke WD, Ding Y (2011) Development and characterization of microsatellite markers for Sagittaria trifolia var. sinensis (Alismataceae). Am J Bot 98:e36–e38
Xu Y, Ma R-C, Xie H, Liu J-T, Cao M-Q (2004) Development of SSR markers for the phylogenetic analysis of almond trees from China and the Mediterranean region. Genome 47:1091–1104
Xu J, Luo X, Zhao D (2012) Cloning and expression of starch synthesis related enzyme gene in common cassava Biote/chnology Bulletin, (11) 101:109
Yakimowski SB, Rymer PD, Stone H, Barrett SC, Dorken ME (2009) Isolation and characterization of 11 microsatellite markers from Sagittaria latifolia (Alismataceae). Mol Ecol Resour 9:579–581
Ye J et al (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
You Y, Liu D, Liu H, Zheng X, Diao Y, Huang X, Hu Z (2015) Development and characterisation of EST-SSR markers by transcriptome sequencing in taro (Colocasia esculenta (L.) Schoot). Mol Breed 35:134
Yue X, Chen J, Guo Y, Wang Q (2011) Population genetic structure of Sagittaria natans (Alismataceae), an endangered species in China, revealed by nuclear SSR loci analyses. Biochemical Systematics and Ecology 39:412–418
Zeng S, Xiao G, Guo J, Fei Z, Xu Y, Roe BA, Wang Y (2010) Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) maxim. BMC Genomics 11:94
Zhao S-Y, Guo Y, Wang Q-F, Chen J, Liu F, Gituru WR (2017) The extent of clonality and genetic diversity in Sagittaria lichuanensis (Alismataceae), an endemic marsh herb in China
Zheng X, Pan C, Diao Y, You Y, Yang C, Hu Z (2013) Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC genomics 14:490
Acknowledgements
We thank to the editor and reviewer for their constructive suggestions and comments. This research was supported by National Key Technologies R&D Program (No. 2012BAD27B01).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Zhi-Liang Zheng
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Supplementary file 1
– Primer sequences for SSR loci. SSR primers were designed using Primer 3.0. The sequence ID, SSR type, SSR motif type, motif size, location of the SSR, location of primers, primer sequences, length of primers, melting temperature, and product size are provided for each locus. (XLS 682 kb)
Supplementary file 2
– Characterization of 100 primer pairs amplification in 79 individual plants. Primer name, sequence ID, repeat motif, sequence of primers, melting temperature and expected size (bp) are indicated. (XLS 45 kb)
Supplementary file 3
– Details of 79 validated EST-SSR markers and predicted functions of their genes, based on BLASTX. Primer name, sequence ID, repeat motif, expected size (bp), objective size (bp), number of varieties from which it was successfully amplified, N. alleles, PIC value, NR_Function, and KOG_Function are indicated. (XLSX 21 kb)
Rights and permissions
About this article
Cite this article
You, Y., Huang, X., Liu, H. et al. Leaf Transcriptome Analysis and Development of EST-SSR Markers in Arrowhead (Sagittaria trifolia L. Var. Sinensis). Tropical Plant Biol. 13, 189–200 (2020). https://doi.org/10.1007/s12042-019-09242-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-019-09242-2