Abstract
Background
Calanthe masuca and C. sinica are two genetically closely related species in Orchidaceae. C. masuca is widely distributed in Asia, whereas C. sinica is restricted to Yunnan and Guangxi Provinces in southwest China. Both play important roles in horticulture and are under the pressure of population decline. Understanding their genetic background can greatly help us develop effective conservation strategies for these species. Simple sequence repeats (SSRs) are useful for genetic diversity analysis, presumably providing key information for the study and preservation of the wild populations of the two species we are interested in.
Results
In this study, we performed RNA-seq analysis on the leaves of C. masuca and C. sinica, obtaining 40,916 and 71,618 unigenes for each species, respectively. In total, 2,019/3,865 primer pairs were successfully designed from 3,764/7,189 putative SSRs, among which 197 polymorphic SSRs were screened out according to orthologous gene pairs. After mononucleotide exclusion, a subset of 129 SSR primers were analysed, and 13 of them were found to have high polymorphism levels. Further analysis demonstrated that they were feasible and effective against C. masuca and C. sinica as well as transferable to another species in Calanthe. Molecular evolutionary analysis revealed functional pathways commonly enriched in unigenes with similar evolutionary rates in the two species, as well as pathways specific to each species, implicating species-specific adaptation. The divergence time between the two closely related species was tentatively determined to be 3.42 ± 1.86 Mya.
Conclusions
We completed and analysed the transcriptomes of C. masuca and C. sinica, assembling large numbers of unigenes and generating effective polymorphic SSR markers. This is the first report of the development of expressed sequence tag (EST)-SSR markers for Calanthe. In addition, our study could enable further genetic diversity analysis and functional and comparative genomic studies on Calanthe.
Similar content being viewed by others
Background
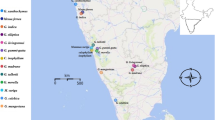
Calanthe R. Br. is a large genus in Orchidaceae, comprising 207 [1] epiphytic and terrestrial species. These species are widely distributed across the pantropical area, and southeast Asia is one of the diversity centres of Calanthe [1]. Subgenus Calanthe section Calanthe series Sylvatica of genus Calanthe includes 14 species widely distributed from southeast Asia to Africa, with only one species, Calanthe sylvatica (Thouars) Lindl., distributed on the African continent. Few studies have focused on Sylvatica species, although these species are of high horticultural value and play an important role in breeding. Two of these species are from China – the widely distributed Calanthe masuca (D. Don) Lindl. and the narrowly distributed C. sinica Z. H. Tsi (Fig. 1), which is restricted to southwest China. A molecular phylogeny based on both nuclear and cpDNA markers showed that these two species are genetically closely related [2]. These two species have the same number of chromosomes (2n = 40) [3, 4] and perhaps large genomes (the genus average C-value is ~ 11.23 pg) [5]. C. sinica is assessed as CR (critically endangered) according to IUCN Red List Categories and Criteria. Recent years have seen rapid population decline for these species, likely due to destruction of habitat and collection. Therefore, research on these species is necessary for further protection strategies. With these closely related species, it is possible to investigate the relationship between Calanthe species from Asia and Africa as well as the driving force underlying the divergence between Sylvatica species.
Molecular markers are important genetic tools for population genetic studies. With advantages of hypervariability, multi-allelic nature, co-dominant inheritance, reproducibility, wide genomic distribution, chromosome-specific location, amenability to automation and high-throughput genotyping [6], simple sequence repeats (SSRs) play an important role in plant genetics and breeding. Currently, next-generation sequencing (NGS) technology provides a convenient and easy way to obtain large-scale EST-SSRs from RNA-sequencing (RNA-seq). SSRs can be predicted after the gain of transcripts from NGS methods [7,8,9]. This technique has been widely used in plants such as Myrica rubra, Ocimum, Vigna angularis [10,11,12] and Orchidaceae [13, 14]. However, the normal method of screening for polymorphism by random selection of SSRs after they have been designated is expensive and time consuming. To solve this problem, Vukosavljev et al. [15] provided a convenient way to screen reads from multiple genotypes for repeats that showed the most length variants, which were subsequently developed into markers rather than simply presented in unfiltered results. This method was effective.
In this study, transcriptome sequencing, de novo assembly and gene annotation were performed on C. masuca and C. sinica, followed by searching for SSRs based on pairwise orthologue relationships, with the purpose of (1) screening out some polymorphic SSRs for further study of C. masuca, C. sylvatica, C. sinica and other species (2). The results of this study will provide transcriptome information for further functional comparative genomics research and SSR-based genetic linkage mapping in Calanthe.
Methods
Materials, total RNA extraction and sequencing
Leaves of Calanthe masuca and C. sinica grown in the nursery of Shanghai Chenshan Botanical Garden were used as the materials for RNA extraction and further analysis. The cultivation and sampling times of the two species were the same to ensure reliable comparability. Further treatments were carried out by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). Young and mature leaves from each species were collected in liquid nitrogen, then total RNA of was isolated using a Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions, and equal amounts of extracted RNA were pooled together. A cDNA library for each RNA-seq was obtained using a TruSeqTM RNA sample prep Kit (Illumina Inc., San Diego, CA, USA) following the manufacturer’s instructions. Subsequently, the cDNA was sequenced using a HiSeq 4000 (Illumina, San Diego, CA, USA) to obtain sequences of ~ 150 bp from both ends of each cDNA.
Illumina sequencing, data filtering, de novo assembly and unigene annotation
Raw reads generated by the Illumina HiSeq platform (submitted to the NCBI database under accessions SRR5478842 and SRR5478843) were first processed to obtain clean reads by trimming off adaptor-ligated regions and low-quality bases (Q < 20) using SeqPrep [16] and Sickle [17]. De novo transcriptome assembly was performed based on cleaned and qualified reads using the short-read assembly program Trinity [18] with default parameters. The longest transcripts were defined as unigenes. Putative unigenes were compared against the NCBI protein non-redundant (NR) protein database and the Swiss-Prot, Pfam and KEGG databases with an E-value cut-off of 1E-5 using BLASTx (BLAST + 2.2.25 package) [19]. Gene ontology (GO) annotations describing biological processes, molecular functions and cellular components for each unigene were obtained by carrying out the program Blast2GO v.2.6.0 [20]. Kobas [21] was recruited to perform the KEGG metabolic pathway annotation.
Orthologous gene identification and evolutionary rate calculation
Orthologous gene pairs between Calanthe masuca and C. sinica used for primer screening were identified by the method of reciprocal best hits (RBH) based on the results of the Basic Local Alignment Search Tool (BLAST) [22,23,24,25]. As the two species were phylogenetically closely related, nucleotide sequences instead of peptide sequences of the unigenes were used to run a BLAST search to improve accuracy. A python script was developed to retrieve the RBH from the results of reciprocal BLASTn (BLAST+ v 2.2.25 package), for which the cutoff was set as e-value = 1E-5.
Orthologous gene groups were defined using the program Inparanoid with default settings (version 4.1; [26]). Phalaenopsis equestris (Schauer) Rchb. f. was used as the outgroup species [27]. The rate of non-synonymous substitutions per non-synonymous substitution site (Ka), the rate of synonymous substitutions per synonymous substitution site (Ks), and the ratio of non-synonymous to synonymous substitution rates (Ka/Ks) for each orthologous gene pair between C. masuca and C. sinica were calculated using the method ‘GMYN’ implemented in the toolbox KaKs_Calculator (version 2.0; [28]). To calculate the Ka and Ks values, the peptide sequences of each pair of orthologous genes were first aligned using the program MAFFT [29] with default settings and were then back-translated into nucleotide sequence alignment using the program PAL2NAL [30].
GO enrichment analysis
GO enrichment analysis was performed using the R package topGO [31], with Fisher’s exact test as the method of enrichment evaluation, all unigenes with GO annotation as the whole list of background genes, and other parameters as default.
EST-SSR markers validation
The software Msatcommander [7] was run on the assembled transcript sequences to find SSR sites and generate primers. The unigenes of two transcriptomes were used to run Msatcommander with primer sets subsequently designed automatically. SSR loci with variations between two species were then screened out in two steps. First, sequence pairs containing SSRs were extracted from the Msatcommander results based on one-to-one correspondence by RBH search. Second, sequences containing the same SSR type (including reverse complement repeat sequences) but with different repeat numbers were selected by comparing the information of microsatellite repeats for each pair of orthologs. The primers for mononucleotide SSRs were then excluded. We only took the primer list of C. masuca as a result by considering that primers identified from two orthologous unigenes will yield the same product.
Development and evaluation of EST-SSR markers
Genomic DNA was isolated from silica-gel-dried leaves using the DNAsecure Plant kit (TIANGEN Inc. Beijing, China) following the manufacturer’s protocol and stored in TE buffer (pH = 8.0). DNA purity and concentration were measured with a NanoDrop 2000c UV-Vis spectrophotometer (Thermo Fisher Scientific Inc., USA). DNA was adjusted to a final concentration of 30 ng/ml and stored at − 20 °C until use.
The primers of the selected SSR alleles were synthesized and amplified, and an M13 (− 21) sequence was added to the forward primers of these primers. An additional primer labelled with fluorescent dye (5′FAM, 5′HEX, or 5′ROX), M13 (− 21), was also synthesized. All primers were synthesized by Sangon Biological Engineering Technology & Service Co. (Shanghai, China). Amplification was carried out in a Thermocycler Mastercycler Gradient (Eppendorf, Germany) using a 20 μl reaction system containing 10 μl of 2× Taq PCR Master Mix (TIANGEN Inc. Beijing, China), 8 μl of ddH2O, 0.2 μl of forward primer (10 μM), 0.4 μl of reverse primer (10 μM) and 0.4 μl of fluorescent dye-labelled M13 primer (10 μM), and 1 μl of target DNA template (0.5 ng/μl). First, PCR was performed for every pair of primers to determine the optimal annealing temperature following Schuelke [32]: 5 min denaturation at 94 °C; 30 cycles of 30 s at 94 °C, 45 s at 53–65 °C, and 45 s at 72 °C; 8 cycles of 30 s at 94 °C, 45 s at 53 °C, and 45 s at 72 °C; a final 10 min extension at 72 °C; and 4 °C hold for storage. After determining the optimal annealing temperature of each primer, amplification was performed.
The PCR products were mixed according to the different fluorescence colour (6-FAM:HEX:ROX = 1:1:1) and then analysed on an ABI 3730 with internal size standard GS500 (GeneScanTM, Applied Biosystems, USA). Allele binning and calling were conducted using Software GeneMarker® V2.2.0 (SoftGenetics, USA). The number of alleles (Na), observed heterozygosity (Ho), gene diversity (expected heterozygosity; He), and polymorphism information content (PIC) for each of the EST-SSR markers were calculated using Cervus software [33].
Before screening for polymorphisms of SSR primers in multiple individuals, all selected primers were first amplified using a single sample, JX1 (C. masuca), and only well-amplified SSR primers were used for further testing. Then, we selected eight individuals from each population collected from Yunnan, Jiangxi, Napo, and Guangxi in China (Additional file 1) for polymorphic analysis. Finally, we chose primers with high PIC values to amplify 72 other individuals (24 from Jiangxi, 2 from Napo, 11 from Yunnan, 15 from Taiwan and 20 from Jinxiu Guangxi). In total, 96 accessions from five populations of three species were used for genetic diversity analysis, including three Calanthe masuca populations (JX, GXJX, TW), one C. sinica population (GXNP) and one C. triplicata population (YN) (Additional file 1).
Universality evaluation of SSR primers
The universality of the developed SSR primers in Calanthe was calculated by in silico PCR. The SSR primers of C. masuca and C. sinica were searched against the unigenes of each other and against the genome sequences of P. equestris [27] and Dendrobium catenatum Lindl. [34]. e-PCR [35] version 2.3.9 was carried out with the SSR primers as queries and the unigenes as databases with no mismatch allowed.
Results
De novo sequence assembly and orthologous genes
In total, 96,697,776/84,001,406 (C. masuca/C. sinica) reads with a length of 151 bp were obtained from the Illumina HiSeq sequencing platform, and 94,826,358/82,290,826 clean reads for the two species, respectively, were obtained after filtering out low-quality reads and adaptors. Using the de novo assembly tool Trinity, we obtained 40,916/71,618 unigenes with an average length of 703.67/625.38 bp, and the N50 values were 1,196/1,086 bp, respectively (Table 1). With the RBH method, 25,152 pairs of putative orthologous genes between the two species were defined (Additional file 2). The distributions of unigene size for both species are shown in Fig. 2. The GC contents for C. masuca and C. sinica unigenes were 44.44%/43.06%, respectively.
Functional annotation and classification
All unigenes of both species were annotated against five databases (Pfam, KEGG, String, Swiss-Prot and NR databases) using BLASTx alignment with an E-value threshold of 1E-5 (Table 2). In total, 40,916/71,618 unigenes were obtained in C. masuca and C. sinica, of which 23,932 (58.5%)/28,037 (39.1%) unigenes of the two species, respectively, returned values when searched against the NR nucleotide database. E-values and similarity distributions of unigenes annotated in the NR database can be found in Table 3. The top three species against which BlastX hits could be identified for unigenes of the two Calanthe species were Phoenix dactylifera (13360, 32.65%/13151, 18.36%) (unigenes number, the percentage of the hit unigenes), Vitis vinifera (1422, 3.48%/2389, 3.34%) and Oryza sativa (590, 1.44%/796, 1.11%) (see Additional file 3 for matched species).
COG and GO classification
By sequence comparison with genes from COG (Clusters of Orthologous Groups of proteins), we found that 9,851 unigenes (24.1% out of all unigenes) of C. masuca and 11,088 unigenes (15.5%) of C. sinica, respectively, could be classified into 26 categories (Fig. 3). The largest groups for both species were assigned to the cluster of ‘General function prediction’ (782, 1.91%/838, 1.17%), followed by ‘Signal transduction mechanisms’ (663, 1.62%/768, 1.07%), ‘Posttranslational modification, protein turnover, chaperones’ (593, 1.45%/624, 0.87%), ‘Translation, ribosomal structure and biogenesis’ (528, 1.29%/615, 0.86%), and ‘Carbohydrate transport and metabolism’ (344, 0.84%/385, 0.54%). The smallest sets were ‘Cell motility’ (6, 0.01%/4, 0%), ‘Nuclear structure’ (1, 0%/1, 0%) and ‘Extracellular structures’ (0, 0%/0, 0%).
We then mapped our unigenes of the two species onto the annotation terms of the three categories in the GO classification system, i.e., biological processes, cellular components, and molecular function. A total of 12,042 (29.43%)/14,044 (19.61%) unigenes in C. masuca and C. sinica, respectively, were annotated to have at least one GO term (Fig. 4). The results showed a high degree of consistency in GO analysis between the two species. The top three groups with most gene members in the category of biological process were metabolic processes (7,509, 18.35%/8,879, 12.40%), cellular processes (6,890, 16.84%/8,198, 11.45%) and single-organism processes (5,121, 12.52%/5,407, 7.55%); the top three largest groups in the category of cellular components were cell (5,609, 13.71%/6,031, 8.42%), cell parts (5,609, 13.71%/6,031, 8.42%), and organelles (4,388, 10.72%/4,736 6.61%). We annotated most genes in both species with molecular functions of binding (6,312, 14.65%/8,016, 11.19%), catalytic activity (5,996, 14.65%/6,763, 9.44%), and transporter activity (726, 1.77%/788, 1.10%).
A preliminary investigation into the evolutionary pattern of genes in Calanthe masuca and C. sinica
With the transcriptome data of two closely related Calanthe species available, it is possible for us to investigate the conservation and divergence pattern of genes in this genus, especially in the lineage represented by C. masuca and C. sinica. To more accurately define orthologous gene pairs, we employed the software Inparanoid (version 4.1; [26]) to define orthologous gene groups between the two species studied here, using P. equestris as the outgroup species. We then calculated the Ks, Ka, and Ka/Ks for each possible orthologous gene pair between C. masuca and C. sinica. We obtained the Ka, Ks, and Ka/Ks values for 10,329 orthologous gene groups between the two species, comprising 12,508 C. masuca unigenes and 11,493 C. sinica unigenes. The Ka/Ks value measures the average selective pressure of the two compared protein-coding sequences since their divergence. To gain preliminary insights into the distinct evolutionary pattern of genes involved in different functional pathways in Calanthe, we divided the genes in each species into 5 different groups according to their Ka/Ks values, namely, the group with strong purifying selection (Ka/Ks ≤ 0.5, abbreviated as SP), the group with intermediately purifying selection (0.5 < Ka/Ks ≤ 0.9, IP), the group with relaxed or nearly relaxed selection (0.9 < Ka/Ks ≤ 1.1, R), the group with intermediately positive or Darwinian selection (1.1 < Ka/Ks ≤ 1.5, ID), and the group with strong Darwinian selection (Ka/Ks > 1.5, SD).
The top enriched GO terms in each corresponding group were quite consistent between the two species (Additional file 4). For example, in the SP group of genes in both species, the biological process ‘oxidation-reduction process’ (GO: 0055114) stood out as the most enriched GO term, with 348 and 332 associated unigenes in C. masuca and C. sinica, respectively. Similarly, ‘folic acid-containing compound biosynthetic process’ and ‘phosphate ion transport’ were among the top 3 enriched GO terms in the IP group in both species. A systematic comparison demonstrated that, in most cases, the shared enriched GO terms between corresponding gene groups of the two species outnumbered GO terms specifically enriched in each species (Table 4), which further validated the assembly and annotation of our RNA-seq data. Notably, the biological process ‘aldehyde biosynthetic process’ (GO: 0046184) was among the top three enriched GO terms in both species, implying fast-evolving indirect plant defence-responsive factors that could help Calanthe species adapt to local environments [36].
To understand the above evolutionary pattern in a real time-scale of species evolution, we set to estimate the divergence time between the two closely related Calanthe species. For a more robust estimation, we determined the minimum Ks values for each of 10,329 orthologous gene groups and determined the mode of the distribution of these Ks values at ~ 0.0328 (Fig. 5). Using the same method, we obtained orthologous gene groups between two epidendroid orchid species, i.e., P. equestris and D. catenatum, and calculated the Ks values for each orthologue pair. The mode of distribution of the minimum Ks values of the orthologous gene groups between P. equestris and D. catenatum was determined at 0.413 (Fig. 5). With the divergence time between these two epidendroid orchids estimated at ~ 43 Mya [37] and assuming that the non-synonymous substitution rates between the two epidendroid and the two Calanthe species were close, we proposed that C. masuca and C. sinica diverged from each other approximately 3.42 ± 1.86 Mya. This preliminary estimation of the divergence time between C. masuca and C. sinica could be improved in the future with more data and information available.
Determination of divergence time between C. masuca and C. sinica. The distribution of minimal synonymous substitutions per synonymous substation sites (Ks) of the orthologous gene groups between C. masuca and C. sinica (a) and between Phalaenopsis equestris and Dendrobium catenatum (b). The divergence time between P. equestris and D. catenatum determined in the publication [37] was used as real-time scale proportional to the measure of Ks
SSR identification, screening of variations and validation
In total, 3,764 putative SSRs in C. masuca and 7,189 SSRs in C. sinica were identified. The majority of the SSRs in both species were either dinucleotides (1,137 in C. sinica and 2,058 in C. masuca) or trinucleotides (1,118 in C. sinica and 1,714 in C. masuca), with longer motifs accounting for only a small proportion (Fig. 6). Finally, 2,019/3,865 primer pairs were successfully designated by Msatcommander for the two species (Additional file 5). In these primers, we screened 197 polymorphic SSR loci according to 25,152 pairs of putative orthologous genes identified by the method of reciprocal best hit (Additional file 5). In addition, 129 SSR loci remained after mononucleotides were excluded (Additional file 5).
Validation of SSR markers
To test the amplification quality and the polymorphisms for the preliminary analysis of the 129 SSR markers, a single sample, JX1 (C. masuca), was used, and it was found that 75 SSRs resulted in clear peaks with little disturbance.
We then performed polymorphic analysis using eight individuals from each population collected from Yunnan, Jiangxi, Napo, and Guangxi in China (Additional file 1). We found that 73 of the 75 examined loci were polymorphic (97.6%) (Fig. 7 and Additional file 6). The two monomorphic loci regarding these 8 individuals could represent polymorphic sites of the subpopulations from which we collected the samples for RNA sequencing. Although potentially polymorphic for other uninspected populations, the two primers were excluded from further analysis for convenience. Nonetheless, 44 out of the 73 loci were found to have a relatively high polymorphic level (PIC≥0.5) and could thus be used in population genetic studies.
To validate the reliability of the primers of the identified polymorphic loci, 13 primers with high PIC values (PIC≥0.5) were randomly selected to amplify a large number of populations and individuals (other 72 individuals). We found that all primers generated high PIC (PIC≥0.5) values (Additional file 7), confirming that the primers could be used in future genetic analysis of Calanthe.
Universality evaluation of SSR primers
The polymorphic SSR markers screening presented above was performed by comparing the transcriptomes of individuals from subpopulations in the two Calanthe species; henceforth, a limited number of polymorphic SSR loci were identified (Additional file 8). To develop a larger number of candidate polymorphic SSR loci, we performed in silico PCR analysis. For the two Calanthe species, the identified primer pairs were compared between them, and both were compared against two orchids with published genomic sequences, P. equestris [27] and D. catenatum [34]. For stringency, all hits with mismatches against the two non-Calanthe orchids were excluded. As a result, among all the 2,019 C. masuca primer pairs, we identified 1,156 (57.3%) pairs that could be used for C. sinica, and in contrast, only 13 (0.6%) primers for D. catenatum were identified. Similarly, among the 3,865 primers of C. sinica, 869 (22.5%) could be used for C. masuca, and only 14 (0.4%) could be used for D. catenatum. We failed to identify any primers from the two Calanthe species for P. equestris. The matching rate of in silico PCR effectively reflects the genetic relationship of these species, as well as the number of transcripts recovered by our RNA-seq analysis.
Discussion
Calanthe masuca and C. sinica are orchid species that play important roles in horticulture. For these two species, with genome size of about 11.23 pg whole genome sequencing is difficult in short timescales. Transcriptome sequencing is hence an important tool for molecular marker identification and gene discovery [38]. In this study,we sequenced the transcriptome of C. masuca and C. sinica, to identify useful SSR markers and obtain transcriptomic information for further studies. Plant genomic analyses can be performed using NGS technologies to determine whether plants have a complete genome sequence. NGS has been used for de novo transcriptome sequencing and assembly in many organisms, including orchids [39,40,41,42]. In this study, 11 Gbp of data were obtained from leaf tissue of each species and assembled into unigenes.
To a certain extent, the length of the assembly sequence reflected the quality of the transcriptome obtained from the Illumina platform [43]. The average unigene length in this study was shorter than in Phalaenopsis aphrodite (875 bp) but longer than in Cymbidium sinensis (612 bp) [41, 42]. This means that the transcriptome data for Calanthe in this study was assembled efficiently.
The conservation and divergence of Calanthe masuca and C. sinica genes
A previous phylogenetic study suggested that C. masuca and C. sinica were closely related to each other [1, 2]. They are winthin sect. Calanthe series sylvatica. C. masuca and C. sinica are the only two species in this series existing in China. They have many common phenotypic characteristics such as long spur, warty calli on the labellum and similar shape of lip midlobe, but still exhibit many phenotypic differences, such as shape of spur. The GO enrichment analysis carried out for each class of genes separately gave similar results in most cases, i.e., We observed much overlap in the enriched GO terms between the corresponding gene classes of the two species. However, we also noted interesting clues in species-specifically enriched GO terms. The number of specifically enriched GO terms of each category in the ID group of genes in C. sinica was also significantly greater than that in C. masuca, further supporting the possibility of specific local adaptation for C. sinica after its divergence with C. masuca. These distinct evolutionary patterns could have occurred on a relatively short time scale, within ~ 3.42 million years. These observations could be validated and/or broadened in future bio-geographical and functional studies.
EST-SSR prediction and validation
SSRs are tandem repeats of short nucleotide motifs with polymorphisms of a certain length that are spread throughout the genome [44]. EST-SSRs have many intrinsic advantages over genomic SSR markers due to their higher transferability among related species [45]. EST-SSR frequency is dependent on several factors, such as genome structure or composition, arithmetical method for SSR detection, and the parameters for exploration of microsatellites [38]. In the two transcriptomes generated in this study, 3,764/7,189 putative SSRs were predicted that differed in SSR number. This may be the cause of differences in genome structure or composition. In both transcriptomes, AG/CT had the highest frequency, as shown in other species [38, 39, 46], and CG/CG had the lowest frequency. In this study, we performed RBH to screen out 25,152 pairs of putative orthologous genes. After searching for SSR loci among these genes, SSR loci with differences between the two transcriptomes were manually identified. This method resulted in SSR loci differing in at least two individuals, which ensured the development of SSR markers with a high level of success. However, the selected alleles may not truly be polymorphic due to the possibility of errors in orthologue prediction. This is why there were two monomorphic loci in this experiment. Even so, this processing can greatly improve the efficiency of screening polymorphic SSR loci.
In this study, we identified 44 loci with high PIC values, and 13 polymorphic SSR loci were randomly chosen for testing from 96 Calanthe individuals, demonstrating their reliability. The remaining 34 loci may also be reliable. These loci can be used in further studies, such as for investigating genetic linkage mapping, germplasm characterization, fingerprinting, and genetic diversity.
Universality evaluation of SSR primers
Using in silico PCR against genome sequences is a practical method for preselecting primers from transcriptomes because the existence of a large intron between primer pairs will greatly affect the performance of PCR amplification [39]. However, this analysis resulted in no identical SSR loci between P. equestris and Calanthe, possibly because of their distant relationship. D. catenatum had 13/14 hits with C. masuca/C. sinica. If genome data from other species closely related to Calanthe were sequenced and included in the analysis, more hits would be identified.
Conclusions
The RNA-seq and de novo transcriptome assembly of two closely related Calanthe species, C. masuca and C. sinica, was conducted. 13 randomly selected primers with a high polymorphism level generated a high level of PIC. A divergence time of 3.42 ± 1.86 Mya was determined between the two species according to Ks. In the future, genetic diversity studies can be conducted in the two Calanthe species and even other closely related species in this genus. Hence some protection strategy can be developed to save these species.
Abbreviations
- e-PCR:
-
Electronic PCR
- EST-SSR:
-
Expressed sequence tag-SSR
- He :
-
Expected heterozygosity
- Ho :
-
Observed heterozygosity
- Na :
-
Number of alleles
- PIC:
-
Polymorphism information content
- RBH:
-
Reciprocal best hits algorithm
- SSR:
-
Polymorphic microsatellite marker
References
Clayton D, Cribb P. The Genus Calanthe. Borneo: Natural History Publications; 2013.
Zhai JW, Zhang GQ, Li L, Wang M, Chen LJ, Chung SW, et al. A new phylogenetic analysis sheds new light on the relationships in the Calanthe alliance (Orchidaceae) in China. Mol Phylogenet Evol. 2014;77:216–22.
Tanaka R, Karasawa K, Ishida G. Karyomorphological observations on Calanthe of Japan. Bull Hiroshima Bot Gard. 1981:9–62.
Karasawa K, Ishida G. The genus calanthe. Tokyo: Yasaka Shobo (in Japanese); 1998.
Leitch IJ, Kahandawala I, Suda J, Hanson L, Ingrouille MJ, Chase MW, et al. Genome size diversity in orchids: consequences and evolution. Ann Bot. 2009;104:469–81.
Parida SK, Kalia SK, Kaul S, Dalal V, Hemaprabha G, Selvi A, et al. Informative genomic microsatellite markers for efficient genotyping applications in sugarcane. Theor Appl Genet. 2009;118:327–38.
Faircloth BC. Msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour. 2008;8:92–4.
Tang J, Baldwin SJ, Jacobs JM, van der Linden CG, Voorrips RE, Leunissen JA, et al. Large-scale identification of polymorphic microsatellites using an in silico approach. BMC Bioinformatics. 2008;9:374.
Castoe TA, Poole AW, de Koning AJ, Jones KL, Tomback DF, Oyler-McCance SJ, et al. Rapid microsatellite identification from Illumina paired-end genomic sequencing in two birds and a snake. PLoS One. 2012;7:e30953.
Chen H, Liu L, Wang L, Wang S, Somta P, Cheng X. Development and validation of EST-SSR markers from the transcriptome of adzuki bean (Vigna angularis). PLoS One. 2015;10:e0131939.
Jiao Y, Jia HM, Li XW, Chai ML, Jia HJ, Chen Z, et al. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics. 2012;13:201.
Mahajan V, Rather IA, Awasthi P, Anand R, Gairola S, Meena SR, et al. Development of chemical and EST-SSR markers for Ocimum genus. Ind Crop Prod. 2015;63:65–70.
Lu JJ, Suo NN, Hu X, Wang S, Liu JJ, Wang HZ. Development and characterization of 110 novel EST-SSR markers for Dendrobium officinale (Orchidaceae). Am J Bot. 2012;99:e415–20.
Jantasuriyarat C, Ritchuay S, Pattarawat P, Huehnea PS, Sureeporn KN. Development and transferability of EST-SSR and transferability of genomic SSR markers for genetic diversity assessment of Doritis. Biochem Syst Ecol. 2012;45:57–65.
Vukosavljev M, Esselink GD, van 't Westende WP, Cox P, Visser RG, Arens P, et al. Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Mol Ecol Resour. 2015;15:17–27.
John JS. SeqPrep. Available: https://github.com/jstjohn/SeqPrep. 2011.
Joshi N, Fass J. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33)[Software]. 2011.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421.
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6.
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–25.
Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2007;24:319–24.
O'brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–80.
Cai J, Liu X, Vanneste K, Proost S, Tsai WC, Liu KW, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 2015;47:65–72.
Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–12.
Alexa A, Rahnenfuhrer J topGO: enrichment analysis for gene ontology R package version 2. 2010.
Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18:233–4.
Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–106.
Zhang GQ, Xu Q, Bian C, Tsai WC, Yeh CM, Liu KW, et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci Rep-UK. 2016;6:19029.
Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7:541–50.
Chehab EW, Kaspi R, Savchenko T, Rowe H, Negre-Zakharov F, Kliebenstein D, et al. Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS One. 2008;3:e1904.
Zhang GQ, Liu KW, Li Z, Lohaus R, Hsiao YY, Niu SC, et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–83.
Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, et al. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics. 2011;12:451.
An M, Deng M, Zheng S, Song Y. De novo transcriptome assembly and development of SSR markers of oaks Quercus austrocochinchinensis and Q. kerrii (Fagaceae). Ree Genet Genomes. 2016;12:103.
De Paolo S, Salvemini M, Gaudio L, Aceto S. De novo transcriptome assembly from inflorescence of Orchis italica: analysis of coding and non-coding transcripts. PLoS One. 2014;9:e102155.
Zhang J, Wu K, Zeng S, Teixeira da Silva JA, Zhao X, Tian CE, et al. Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral development. BMC Genomics. 2013;14:279.
Su C, Chao Y, Chang A, Chen W, Chen C, Lee A, et al. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011;52(9):1501–14.
Chen X, Xie YH Sun XM. Development and characterization of polymorphic genic-SSR markers in Larix kaempferi. Molecules. 2015;20:6060–7.
Zhou X, Dong Y, Zhao J, Huang L, Ren X, Chen Y, et al. Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.). BMC Genomics. 2016;17:420.
Kong Q, Xiang C, Yu Z. Development of EST-SSRs in Cucumis sativus from sequence database. Mol Ecol Notes. 2006;6:1234–6.
Zhang H, Wei L, Miao H, Zhang T, Wang C. Development and validation of genic-SSR markers in sesame by RNA-seq. BMC Genomics. 2012;13:316.
Acknowledgements
The authors are grateful to Ziyi Ni and Li Shao of Shanghai Chenshan Botanical Garden for their help in caring for the materials in the greenhouse. We all thank Miao An for the help in the data analysis.
Availability of data and material
The datasets generated during the current study are available in Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/) with accession SRR5478842 and SRR5478843.
Funding
This work is supported by the Sino-Africa Joint Center research project (Grant No. SAJC201607 to Weichang Huang) and Shanghai Municipal Administration of Forestation and City Appearances (Grant No. G152431 to Chao Hu). The author Tungyu Hsieh thanks the Chinese Academy of Sciences (CN) for support (Grant No. 2013TW2SA0003, 2015TW1SA0001 to Tungyu Hsieh).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: CH, HY, KJ, WH, SL. Performed the experiments: CH, KJ. Analysed the data: CH, HY, LW. Contributed reagents/materials/analysis tools: CH, WH, BY, TH. Drafted and revised the paper: CH, HY, WH, KJ, SL, LW, BY, TH. Photos: CH and TH. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The locations of material collected here are neither privately owned lands nor protected areas. No specific permits were required for our research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Materials used in this article. (DOCX 14 kb)
Additional file 2:
Table S2. List of Reciprocal Best Hits of C. masuca and C. sinica. (XLSX 741 kb)
Additional file 3:
Table S3. Proportion of matched unigenes in the NR database. (DOCX 15 kb)
Additional file 4:
Table S4. GO annotations of unigenes with Ka/Ks and the results of following enrichment analysis. (XLSX 100 kb)
Additional file 5:
Table S5. List of SSR primer pairs derived from two species and 129 selected SSR primers. (XLSX 1354 kb)
Additional file 6:
Table S6. The statistical parameters of 75 validated primer pairs using eight individuals of each species. (XLSX 18 kb)
Additional file 7:
Table S7. The statistical parameters of 13 validated primer pairs using 96 individuals. (XLSX 12 kb)
Additional file 8:
Table S8. In silico PCR analysis. Using the primers of C. masuca and C. sinica against the unigene of each other and against Phalaenopsis equestris and Dendrobium catenatum. (XLSX 114 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hu, C., Yang, H., Jiang, K. et al. Development of polymorphic microsatellite markers by using de novo transcriptome assembly of Calanthe masuca and C. sinica (Orchidaceae). BMC Genomics 19, 800 (2018). https://doi.org/10.1186/s12864-018-5161-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-5161-4