Abstract
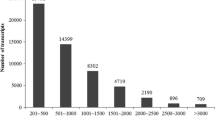
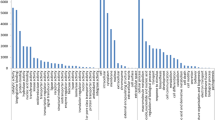
Illumina-based paired-end sequencing technology was used for the high-throughput transcriptome sequencing of combined Zingiber striolatum Diels tissues (i.e., root, stem, leaf, flower, and fruit tissues). More than 130 million sequencing reads were generated, and a de novo assembly yielded 287,959 contigs and 112,107 unigenes with an average length of 1029 and 28,891 bp, respectively. Similarity searches with known sequences led to the identification of 51,804 (46.21%) genes. Of the annotated unigenes, 6867 and 51,987 were assigned to Gene Ontology and Clusters of Orthologous Groups categories, respectively. Additionally, 8384 simple sequence repeats (SSRs) were identified as potential molecular markers in the unigenes. Thirty pairs of polymerase chain reaction primers were designed and used to validate the unigenes and assess the associated genomic polymorphism. The PCR amplification products for 25 primer pairs were of the expected size. These primers may represent usable molecular markers. The thousands of SSR markers identified in the present study may be useful for analyses of genetic diversity, genetic linkage mapping, and the identification and improvement of varieties during the breeding of Z. striolatum Diels. The unigene sequences and SSR markers described herein may serve as valuable resources for future investigations of Z. striolatum Diels.



Similar content being viewed by others
References
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V et al (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114(2):359–372
An editorial committee of flora of China (1981) Zingiber striolatum Diels. Flora of China. Science Press, Beijing, p 146
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D et al (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156(2):847–854
Chen H, Song Y, Li LT, Khan MA, Li XG et al (2014) Construction of a high-density simple sequence repeat consensus genetic map for pear (Pyrus spp.). Plant Mol Biol Rep 33(2):1–10
Cheng JW, Zhao ZC, Li B, Qin C, Wu ZM et al (2016) A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci Rep 6:189–190
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160(6):1115–1123
Ge J, Xie H, Chui CS, Hong JM, Ma RC (2005) Analysis of expressed sequence tags (ESTs) derived SSR markers in Chinese cabbage (Brassica campestris L. ssp. pekinensis). J Agric Biotechnol 13(4):423–428
Goldstein DB, Linares AR, Cavalli-Sforza LL, Feldman MW (1996) An evaluation of genetic distances for use with microsatellite loci. Genetics 139(1):463–471
Jin JQ, Cui HR, Chen WY, Lu MZ, Yao YL et al (2006) Data mining for SSRs in ESTs and development of EST-SSR marker in tea plant (Camellia sinensis). J Tea Sci 26(1):17–23
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD (2008) The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res 18(1):30–38
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998
Li HS, Fan SL, Sheng FF (2005) Screening of microsatellite markers from cotton ESTs. Cotton Sci 17(4):211–216
Liang X, Chen X, Hong Y, Liu H, Zhou G et al (2009) Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol 9:35–48
Liu F, Wang YS, Tian XL, Mao ZC, Zou XX et al (2012) SSR mining in pepper(Capsicum annuum L.)transcriptome and the polymorphism analysis. Acta Hortic Sin 39(1):168–174
Moe AM, Weiblen GD (2011) Development and characterization of microsatellite loci in dioecious figs (Ficus, Moraceae). Am J Bot 98(2):e25–e27
Portis E, Nagy I, Sasvari Z, Stagel A, Barchi L et al (2007) The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci 172:640–648
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1(7):215–222
Provan J, Kumar A, Shepherd L, Powell W, Waugh R (1996) Analysis of intra-specific somatic hybrids of potato (Solanum tuberosum) using simple sequence repeats. Plant Cell Rep 16(3–4):196–199
Qu L, Xia LS, Liu D, Feng WY (2015) Rrogress in Zingiber striolatum Diels. Yunnan J Tradit Chin Medi Mater Med 5:111–113
Rongwen J, Akkaya M, Bhagwat A, Lavi U, Cregan P (1995) The use of microsatellite DNA markers for soybean genotype identification. Theor Appl Genet 90(1):43–48
Tan S, Cheng JW, Zhang L, Qin C, Nong DG et al (2015) Construction of an interspecific genetic map based on InDel and SSR for mapping the QTLs affecting the initiation of flower primordia in pepper (Capsicum spp.). PLoS ONE 10:1–15
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsa tellites in ESTs of some cereal species. Cell Mol Biol Lett 7(2A):537–546
Wang JY, Chen YY, Liu WL, Wu YT (2008) Development and application of EST-derived SSR markers for bananas (Musa nana Lour.). Hereditas 30(7):933–940
Wang HL, Yang J, Boykin LM, Zhao QY, Wang YJ et al (2014) Developing conversed microsatellite markers and their implications in evolutionary analysis of the Bemisia tabaci complex. Sci Rep 4:1–10
Yi G, Lee JM, Lee S, Choi D, Kim BD (2006) Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet 114:113–130
Yu Y, Saghai Maroof M, Buss G, Maughan P, Tolin S (1993) RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology 84(1):60–64
Zalapa JE, Cuevas H, Zhu H, Steffan S, Senalik D et al (2012) Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am J Bot 99:193–208
Zhang QT, Li XY, Yang YM, Fan ST, Ai J (2016) Analysis on SSR information in transcriptome and development of molecular markers in Lonicera caerulea. Acta Hortic Sin 43(3):557–563
Zong XJ, Wang WW, Wang JW, Wei HR, Yan XR et al (2012) The application of SYBR Green I real-time quantitative RT-PCR in quantitative analysis of sweet cherry viruses in different tissues. Acta Phytophylacica Sin 39(6):497–502
Acknowledgements
Professor William Yajima is gratefully acknowledged for correction to the manuscript. This research was supported by the Guizhou innovation talent base construction of potato industry technology ([2016]22), the technical innovation fund for small and medium-sized enterprises funded projects (13C26215205306) and Guizhou science and technology project ([2016]2554).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deng, K., Deng, R., Fan, J. et al. Transcriptome analysis and development of simple sequence repeat (SSR) markers in Zingiber striolatum Diels. Physiol Mol Biol Plants 24, 125–134 (2018). https://doi.org/10.1007/s12298-017-0485-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-017-0485-0