Abstract
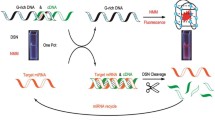
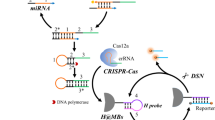
The development of a bio-sensing strategy based on CRISPR/Cas that is exceptionally sensitive is crucial for the identification of trace molecules. Colorimetric miRNA detection utilizing CRISPR/Cas13a-triggered DNAzyme signal amplification was described in this article. The developed strategy was implemented for miRNA-21 detection as a proof of concept. The cleavage activity of Cas13a was triggered when the target molecule bonded to the Cas13a-crRNA complex and cleaved uracil ribonucleotides (rU) in the substrate probe. As a consequence, the S chain was liberated from the T chain that had been modified on magnetic beads (MB). The G-rich sections were then exposed when the catalytic hairpin assembly between the H1 and H2 probes was activated by the released T@MB. G-rich section can fold into G-quadruplex. By catalyzing the formation of green ABTS3– via HRP-mimicking G-quadruplex/hemin complexes, colorimetric measurements of miRNA can be achieved visually through DNAzyme-mediated signal amplification. The method demonstrated a low limit of detection of 27 fM and a high selectivity towards target miRNA eventually. As a result, the developed strategy provides a clinical application platform for the detection of miRNAs that is both ultrasensitive and extremely specific.






Similar content being viewed by others
Data Availability
All data generated and analyzed during this study are included in this article.
References
Nikanjam, M., Kato, S., & Kurzrock, R. (2022). Liquid biopsy: Current technology and clinical applications. Journal of Hematology & Oncology, 15(1), 131.
Zhang, Z., Wu, H., Chong, W., Shang, L., Jing, C., & Li, L. (2022). Liquid biopsy in gastric cancer: Predictive and prognostic biomarkers. Cell Death & Disease, 13(10), 903.
Lin, B., Jiang, J., Jia, J., & Zhou, X. (2022). Recent advances in exosomal miRNA biosensing for liquid biopsy. Molecules, 27(21), 7145.
Markou, A., Tzanikou, E., & Lianidou, E. (2022). The potential of liquid biopsy in the management of cancer patients. Seminars in Cancer Biology, 84, 69–79.
Chen, L., Heikkinen, L., Wang, C., Yang, Y., Sun, H., & Wong, G. (2019). Trends in the development of miRNA bioinformatics tools. Briefings in Bioinformatics, 20(5), 1836–1852.
Mishra, S., Yadav, T., & Rani, V. (2016). Exploring miRNA based approaches in cancer diagnostics and therapeutics. Critical Reviews in Oncology Hematology, 98, 12–23.
Fan, C., Li, Y., Lan, T., Wang, W., Long, Y., & Yu, S. Y. (2022). Microglia secrete miR-146a-5p-containing exosomes to regulate neurogenesis in depression. Molecular Therapy, 30(3), 1300–1314.
Song, M. (2020). miRNAs-dependent regulation of synapse formation and function. Genes Genomics, 42(8), 837–845.
Rahmani, S., Kadkhoda, S., & Ghafouri-Fard, S. (2022). Synaptic plasticity and depression: The role of miRNAs dysregulation. Molecular Biology Reports, 49(10), 9759–9765.
Narayanan, R., & Schratt, G. (2020). miRNA regulation of social and anxiety-related behaviour. Cellular and Molecular Life Sciences, 77(21), 4347–4364.
Torres, A. G., Fabani, M. M., Vigorito, E., & Gait, M. J. (2011). MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA, 17(5), 933–943.
Kim, K. J., Kwak, J., Lee, J. H., & Lee, S. S. (2017). Real-time qRT-PCR assay for the detection of miRNAs using bi-directional extension sequences. Analytical Biochemistry, 536, 32–35.
Li, W., & Ruan, K. (2009). MicroRNA detection by microarray. Analytical and Bioanalytical Chemistry, 394(4), 1117–1124.
Ren, A., Dong, Y., Tsoi, H., & Yu, J. (2015). Detection of miRNA as non-invasive biomarkers of colorectal cancer. International Journal of Molecular Sciences, 16(2), 2810–2823.
Cheng, Y., Dong, L., Zhang, J., Zhao, Y., & Li, Z. (2018). Recent advances in microRNA detection. The Analyst, 143(8), 1758–1774.
Granados-Riveron, J. T., & Aquino-Jarquin, G. (2021). CRISPR/Cas13-Based approaches for ultrasensitive and specific detection of microRNAs. Cells, 10(7), 1655.
Wang, Y. H., He, L. L., Huang, K. J., Chen, Y. X., Wang, S. Y., Liu, Z. H., & Li, D. (2019). Recent advances in nanomaterial-based electrochemical and optical sensing platforms for microRNA assays. The Analyst, 144(9), 2849–2866.
Hosseinzadeh, E., Ravan, H., Mohammadi, A., & Pourghadamyari, H. (2020). Colorimetric detection of miRNA-21 by DNAzyme-coupled branched DNA constructs. Talanta, 216, 120913.
Nie, N., Tang, W., Ding, X., Guo, X., & Chen, Y. (2022). DNAzyme based dual signal amplification strategy for ultrasensitive myocardial ischemia related MiRNA detection. Analytical Biochemistry, 640, 114543.
Wang, Y., Feng, H., Huang, K., Quan, J., Yu, F., Liu, X., Jiang, H., & Wang, X. (2022). Target-triggered hybridization chain reaction for ultrasensitive dual-signal miRNA detection. Biosensors & Bioelectronics, 215, 114572.
Yuan, J. H., Shao, W., Chen, S. B., Huang, Z. S., & Tan, J. H. (2020). Recent advances in fluorescent probes for G-quadruplex nucleic acids. Biochemical and Biophysical Research Communications, 531(1), 18–24.
Xie, Y., Zhang, S., Deng, T., Zhang, K., Ren, J., & Li, J. (2021). A Novel DNAzyme signal amplification-based colorimetric method for RNase H assays. Analytical Sciences, 37(12), 1675–1680.
Shahsavar, K., Shokri, E., & Hosseini, M. (2022). Sensitive colorimetric detection of miRNA-155 via G-quadruplex DNAzyme decorated spherical nucleic acid. Mikrochimica Acta, 189(9), 357.
Asa, T. A., Ravi Kumara, G. S., & Seo, Y. J. (2022). Highly sensitive, selective, and rapid detection of miRNA-21 using an RCA/G-quadruplex/QnDESA probing system. Analytical Methods, 14(2), 97–100.
Zhang, Q., Zhang, X., Zou, X., Ma, F., & Zhang, C. Y. (2023). CRISPR/Cas-based MicroRNA Biosensors. Chemistry (Easton), 29(16), e202203412.
Zhao, D., Tang, J., Tan, Q., Xie, X., Zhao, X., & Xing, D. (2023). CRISPR/Cas13a-triggered Cas12a biosensing method for ultrasensitive and specific miRNA detection. Talanta, 260, 124582.
Funding
The research was financially supported by the Hengshui City Science and Technology Plan Project (project number: 2023014010Z).
Author information
Authors and Affiliations
Contributions
Z.Y. is the supervisor of the team in all research steps including designing, data analysis, and manuscript writing. N.Y. has the main role for experimental data collection, data gathering, preparation of results, and data analysis. Z.H. and L.Z. assisted in the data analysis.
Corresponding author
Ethics declarations
Ethics Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, N., Hu, Z. & Zhang, L. CRISPR-Cas13a-Triggered DNAzyme Signal Amplification-Based Colorimetric miRNA Detection Method and Its Application in Evaluating the Anxiety. Appl Biochem Biotechnol (2024). https://doi.org/10.1007/s12010-024-04951-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-024-04951-1