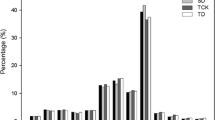
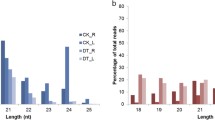
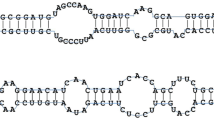
Abstract
MicroRNAs (miRNAs) are a class of small non-coding, single-stranded RNA sequences that regulate gene expression at the post-transcriptional level and also reported to function in stress responses, but their role has not been studied in Camelina (Camelina sativa L.), an emerging oil crop. In this study, we predicted conserved as well as putative novel miRNAs from a Camelina drought stress cDNA library using comprehensive genomic approaches. Based on the sequence homology, we predicted 145 miRNAs, of which 61 were conserved, and 84 putative novel miRNAs were found to belong to 26 and 72 different miRNA families, respectively. In silico expression analysis indicated that 20 miRNAs were really expressed in Camelina genome, and several of them have tissue-specific expression character. We found that the 60 putative novel miRNA families target 117 genes. Most of the miRNA targets were predicted to genes including that regulate stress response, transcription factors, and fatty acid and lipid metabolism-related genes. Expression patterns of 6 randomly selected miRNAs under drought stress were validated by real-time quantitative polymerase chain reaction analysis. Coordinated expression changes between 6 randomly selected miRNAs and their target genes, suggested that the predicted miRNAs could be drought-responsive and that they would likely be directly involved in stress regulatory networks of Camelina. These results indicate that, in C. sativa, under drought stress, a large number of new miRNAs could be discovered, and the predicted stress-responsive miRNAs and their target transcripts will serve as valuable resources for future studies.






Similar content being viewed by others
References
Barciszewska-Pacak M, Milanowska K, Knop K, Bielewicz D, Nuc P, Plewka P, Am Pacak, Vazquez F, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z (2015) Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front Plant Sci 6:410
Bari A, Orazova S, Ivashchenko A (2013) miR156- and miR171-binding sites in the protein-coding sequences of several plant genes. BioMed Res Int 2013:1–7
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. P Natl Acad Sci-Biol USA 102:11928–11933
Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA (2008) Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. Am J Bot 95:1307–1327
Bertolini E, Verelst W, Horner DS, Gianfranceschi L, Piccolo V, Inzé D, Pè ME, Mica E (2013) Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol Plant 6:423–443
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579:5923–5931
Chi X, Yang Q, Chen X, Wang J, Pan L, Chen M, Yang Z, He Y, Liang X, Yu S (2011) Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS ONE 6:e27530
Collins-Silva JE, Lu C, Cahoon EB (2011) Camelina: a designer biotech oilseed crop. Inform 22:610–613
Dhandapani V, Ramchiary N, Paul P, Kim J, Choi SH, Lee J, Hur Y, Lim YP (2011) Identification of potential microRNAs and their targets in Brassica rapa L. Mol Cells 32:1–37
Ding Y, Tao Y, Zhu C (2013) Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot 64:3077–3086
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Felice KM, Salzman DW, Shubert-Coleman J, Jensen KP, Furneaux HM (2009) The 5′ terminal uracil of let-7a is critical for the recruitment of mRNA to Argonaute2. Biochem J 422:329–341
Fujimoto M, Suda Y, Vernhettes S, Nakano A, Ueda T (2015) Phosphatidylinositol 3-kinase and 4-kinase have distinct roles in intracellular trafficking of cellulose synthase complexes in Arabidopsis thaliana. Plant Cell Physiol 56:287–298
Galli V, Guzman F, de Oliveira LF, Loss-Morais G, Körbes AP, Silva SD, Margis-Pinheiro MM, Margis R (2014) Identifying microRNAs and transcript targets in Jatropha seeds. PLoS One 9:e83727
Gehringer A, Friedt W, Luhs W, Snowdon RJ (2006) Genetic mapping of agronomic traits in false flax (Camelina sativa subsp. sativa). Genome 49:1555–1563
Han SK, Sang Y, Rodrigues A, Biol F, Wu MF, Rodriguez PL, Wagner D (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24:4892–4906
Hixson SM, Parrish CC, Anderson DM (2014) Changes in tissue lipid and fatty acid composition of farmed rainbow trout in response to dietary camelina oil as a replacement of fish oil. Lipids 49:97–111
Huang P, Ju HW, Min JH, Zhang X, Kim SH et al (2013) Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci 203–204:98–106
Jian X, Zhang L, Li G, Zhang L, Wang X, Cao X, Fang X, Chen F (2010) Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics 95:47–55
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, Higgins Erin E, Huebert T, Sharpe AG, Parkin IAP (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5:3706
Kantar M, Lucas SJ, Budak H (2011) miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233:471–484
Kanth BK, Kumari S, Choi SH, Ha HJ, Lee GJ (2015) Generation and analysis of expressed sequence tags (ESTs) of Camelina sativa to mine drought stress-responsive genes. Biochem Biophys Res Commun 467:83–93. doi:10.1016/j.bbrc.2015.09.116
Kasukabe Y, He LX, Nada K (2004) Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes transgenic Arabidopsis thaliana. Plant Cell Physiol 45:712–722
Kim HS, Oh JM, Luan S, Carlson JE, Ahn SJ (2013) Cold stress causes rapid but differential changes in properties of plasma membrane H+-ATPase of camelina and rapeseed. J Plant Physiol 170:828–837
Körbes AP, Machado RD, Guzman F, Almerão MP, de Oliveira LF, Loss-Morais G, Turchetto-Zolet AC, Cagliari A, dos Santos Maraschin F, Margis-Pinheiro M, Margis R (2012) Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS ONE 7:e50663
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Kulcheski FR, de Oliveira LF, Molina LG, Almerão MP, Rodrigues FA, Marcolino J, Barbosa JF, Stolf-Moreira R, Nepomuceno AL, Marcelino-Guimarães FC, Abdelnoor RV, Nascimento LC, Carazzolle MF, Pereira GA, Margis R (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom 12:307
Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101:12753–12758
Kwak KJ, Kang H, Han KH, Ahn SJ (2013) Molecular cloning, characterization, and stress-responsive expression of genes encoding glycine-rich RNA-binding proteins in Camelina sativa L. Plant Physiol Biochem 68:44–51
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li XY, Wang C, Nie PP, Lu XW, Wang M, Liu W, Yao J, Liu YG, Zhang QY (2011) Characterization and expression analysis of the SNF2 family genes in response to phytohormones and abiotic stresses in rice. Biol Plant 55:625–633
Li B, Duan H, Li J, Deng XW, Yin W, Xia X (2013) Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol Biol 81:525–539
Liang C, Liu X, Yiu SM, Lim BL (2013) De novo assembly and characterization of Camelina sativa transcriptome by paired-end sequencing. BMC Genom 14:146
Liu H-H, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Liu Z, Kumari S, Zhang L, Zheng Y, Ware D (2012) Characterization of miRNAs in Response to Short-Term Waterlogging in Three Inbred Lines of Zea mays. PLoS One 7:e39786
Liu F, Wang W, Sun X, Liang Z, Wang F (2015) Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ 38:1357–1367
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Marco A, Macpherson JI, Ronshaugen M, Griffiths-Jones S (2012) MicroRNAs from the same precursor have different targeting properties. Silence 3:8
Moser BR (2012) Biodiesel from alternative oilseed feed stocks: camelina and field pennycress. Biofuels 3:193–209
Mudalkar S, Golla R, Ghatty S, Reddy AR (2014) De novo transcriptome analysis of an imminent biofuel crop, Camelina sativa L. using Illumina GAIIX sequencing platform and identification of SSR markers. Plant Mol Biol 84:159–171
Panda D, Dehury B, Sahu J, Barooah ZM, Sen P, Modi MK (2015) Computational identification and characterization of conserved miRNAs and their target genes in garlic (Allium sativum L.) expressed sequence tags. Gene 537:333–342
Poudel S, Aryal N, Lu C (2015) Identification of MicroRNAs and transcript targets in Camelina sativa by deep sequencing and computational methods. PLoS One 10:e0121542
Quah S, Hui JHL, Holland PWH (2015) A burst of miRNA innovation in the early evolution of butterflies and moths. Mol Biol Evol 32:1161–1174
Rajangam AS, Gidda SK, Craddock C, Mullen RT, Dyer JM, Eastmond PJ (2013) Molecular characterization of the fatty alcohol oxidation pathway for wax-ester mobilization in germinated jojoba seeds. Plant Physiol 161:72–80
Rogans SJ, Rey C (2016) Unveiling the micronome of Cassava (Manihot esculenta Crantz). PLoS ONE 11:e0147251
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16:258–264
Séguin-Swartza G, Eyncka C, Gugel RK, Strelkovb SE, Oliviera CY, Lic JL, Klein-Gebbinckd H, Borhana H, Caldwellc CD, Falka KC (2009) Diseases of Camelina sativa (false flax). Can J Plant Pathol 31:375–386
Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11:5
Song ZZ, Yang SY, Zuo J, Su YH (2014) Over-expression of ApKUP3 enhances potassium nutrition and drought tolerance in transgenic rice. Biol Plant 58:649–658
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Taylor RS, Tarver JE, Hiscock SJ, Donoghue PC (2015) Evolutionary history of plant microRNAs. Trends Plant Sci 19:175–182
Theodoulou FL, Holdsworth M, Baker A (2006) Peroxisomal ABC transporters. FEBS Lett 580:1139–1155
Verma SS, Rahman MH, Deyholos MK, Basu U, Kav NN (2014) Differential expression of miRNAs in Brassica napus root following infection with Plasmodiophora brassicae. PLoS One 9:e86648
Wang F, Chen H, Li X, Wang N, Wang T, Yang J, Guan L, Yao N, Du L, Wang Y, Liu X, Chen X, Wang Z, Dong Y, Li H (2015) Mining and identification of polyunsaturated fatty acid synthesis genes active during camelina seed development using 454 pyrosequencing. BMC Plant Biol 15:147
Xie F, Frazier TP, Zhang B (2010) Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta 232:417–434
Xue RG, Zhang B, Xie HF (2007) Overexpression of a NTR1 in transgenic soybean confers tolerance to water stress. Plant Cell Tiss Organ Cult 89:177–183
Xue HW, Chen X, Mei Y (2009) Function and regulation of phospholipid signalling in plants. Biochem J 421:145–156
Yin Z, Li C, Han X, Shen F (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414:60–66
Yu X, Wang H, Lu Y, De Ruiter M, Cariaso M, Prins M, van Tunen A, He Y (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63:1025–1038
Yu G, Li J, Sun X, Zhang X, Liu J, Pan H (2015) Overexpression of AcNIP5;1, a novel nodulin-like intrinsic protein from halophyte Atriplex canescens, enhances sensitivity to salinity and improves drought tolerance in Arabidopsis. Plant Mol Biol Rep 33:1864–1875
Zhang BH, Pan XP, Anderson TA (2006a) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhang BH, Pan XP, Cobb GP, Anderson TA (2006b) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Zhang BH, Pan XP, Stellwag EJ (2008a) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhang Y, Xu W, Li Z, Deng XW, Wu W, Xue Y (2008b) F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol 148:2121–2133
Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, Wang T, Ling Y, Su Z (2010) PMRD: plant microRNA database. Nucleic Acids Res 38:D806–D813
Zhang N, Yang J, Wang Z, Wen Y, Wang J, He W, Liu B, Si H, Wang D (2014) Identification of novel and conserved MicroRNAs related to drought stress in potato by deep sequencing. PLoS One 9:e95489
Zhang SD, Ling LZ, Zhang QF, Xu JD, Cheng L (2015) Evolutionary comparison of two combinatorial regulators of SBP-box genes, MiR156 and MiR529, in plants. PLoS One 10:e0124621
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354:585–590
Zubr J (1997) Oil-seed crop Camelina sativa. Ind Crops Prod 6:113–119
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
This research was supported by Bio-industry Technology Development Program (No. 312033-5), iPET (Korea Insitute of Planning and Evaluation for Technology in Agriculture, Food and Rural Affairs) and Radiation Technology R&D program through the National Research Foundation of Korea funded by the ministry of Science, ICT & Future Planing (NRF-2013M2A2A6043621).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This manuscript has no financial or non-financial competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11816_2016_395_MOESM1_ESM.docx
Procedure of potential Camelina sativa miRNA gene search by identifying homologs of previously known plant miRNAs (DOCX 13 kb)
11816_2016_395_MOESM2_ESM.pdf
Predicted hairpin secondary structures of the 84 putative novel miRNAs identified in this study. Mature miRNA sequences are highlighted in green. The lengths of the accurate miRNA precursors may be slightly longer than that presented here (PDF 1079 kb)
11816_2016_395_MOESM3_ESM.tif
miRNA mir11736–mir3698 cluster in Camelina EST CaSativa1SL019889t001. a Schematic diagram of organization of the cluster. b EST sequence containing miRNAs encoded within the cluster. Underlined sequences indicate the pre-miRNAs; bold red- and green-colored nucleotides represent the mature miRNAs. c The predicted hairpin structures of mir11736 and mir3698 (TIFF 46083 kb)
11816_2016_395_MOESM4_ESM.xlsx
Conserved miRNAs identified by homolog search and secondary structure in the present study. mir: mature miRNA (XLSX 19 kb)
11816_2016_395_MOESM5_ESM.xlsx
Putative novel miRNAs identified by homolog search and secondary structure in the present study. ML: mature sequence length; MS: mature miRNA sequence arm side; LP: length of pre-miRNAs; NM: number of nucleotide mismatches (XLSX 30 kb)
11816_2016_395_MOESM6_ESM.xlsx
In silico blast expression analysis of novel miRNA genes in different tissues and seed developmental stages from the Camelina transcriptome data (Poudel et al. 2015). Number in the columns indicates number of miRNA hits during BLAST as well as expression in that particular tissue (leaves, buds, and seeds). Absence of number describes miRNA with no BLAST hit or absence of expression. On the right side panel, heatmap depicting the tissue-specific transcript accumulation of miRNAs in various tissues which is produced using the number of miRNA hits during BLAST. A gradient color bar scale represents the transcript accumulation level of high (red) or low (green) (XLSX 23 kb)
11816_2016_395_MOESM7_ESM.docx
List of primer sequences used for qRT-PCR analyses in the present study. miRNA-specific forward primers in combination with universal reverse primer from Mir-X™ miRNA First-Strand synthesis kit (Clontech, USA) were used (DOCX 12 kb)
11816_2016_395_MOESM8_ESM.xlsx
Potential targets of putative novel identified Camelina sativa miRNAs and similar Arabidopsis thaliana genes (XLSX 32 kb)
Rights and permissions
About this article
Cite this article
Subburaj, S., Kim, A.Y., Lee, S. et al. Identification of novel stress-induced microRNAs and their targets in Camelina sativa using computational approach. Plant Biotechnol Rep 10, 155–169 (2016). https://doi.org/10.1007/s11816-016-0395-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-016-0395-6