Abstract
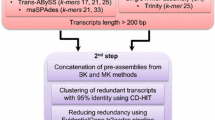
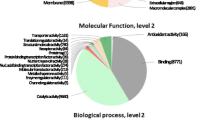
Camelina sativa L. is an emerging biofuel crop with potential applications in industry, medicine, cosmetics and human nutrition. The crop is unexploited owing to very limited availability of transcriptome and genomic data. In order to analyse the various metabolic pathways, we performed de novo assembly of the transcriptome on Illumina GAIIX platform with paired end sequencing for obtaining short reads. The sequencing output generated a FastQ file size of 2.97 GB with 10.83 million reads having a maximum read length of 101 nucleotides. The number of contigs generated was 53,854 with maximum and minimum lengths of 10,086 and 200 nucleotides respectively. These trancripts were annotated using BLAST search against the Aracyc, Swiss-Prot, TrEMBL, gene ontology and clusters of orthologous groups (KOG) databases. The genes involved in lipid metabolism were studied and the transcription factors were identified. Sequence similarity studies of Camelina with the other related organisms indicated the close relatedness of Camelina with Arabidopsis. In addition, bioinformatics analysis revealed the presence of a total of 19,379 simple sequence repeats. This is the first report on Camelina sativa L., where the transcriptome of the entire plant, including seedlings, seed, root, leaves and stem was done. Our data established an excellent resource for gene discovery and provide useful information for functional and comparative genomic studies in this promising biofuel crop.










Similar content being viewed by others
References
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Atabani AE, Silitonga AS, Badruddin Irfan Anjum et al (2012) A comprehensive review on biodiesel as an alternative energy resource and its characteristics. Renew Sust Energ Rev 16(4):211–245
Brown AP, Kroon JTM, Swarbreck D et al (2012) Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS ONE 7(2):e30100
Cheung F, Win J, Lang JM, Hamilton J et al (2008) Analysis of the Pythium ultimum transcriptome using Sanger and Pyrosequencing approaches. BMC Genomics 9:542
Cloutier S, Niu Z, Datla R, Duguid S (2009) Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor Appl Genet 119(1):53–63
Collins LJ, Biggs PJ, Voelckel C, Joly S (2008) An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Inform 21:3–14
Conesa A, Gotz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832
Costa GG, Cardoso KC, Bem Del et al (2010) Transcriptome analysis of the oil-rich seed of the bioenergy crop Jatropha curcas L. BMC Genomics 11(1):462
DiGuistini S, Liao N, Platt D et al (2009) De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol 10(9):R94
Dutta S, Kumawat G, Singh BP et al (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea (Cajanus cajan (L.) Millspaugh). BMC Plant Biol 11:17
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18(1):53–63
Grabherr MG, Haas BJ, Yassour M et al (2011) Full length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hutcheon C, Ditt RF, Beilstein M et al (2010) Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol 10:233
Illumina (2009) mRNA sequencing sample preparation guide. Illumina 24
Kashi Y, King DG (2006) Simple sequence repeats as advantageous mutators in evolution. Trends Genet 22(5):253–259
Kudapa H, Bharti AK, Cannon SB et al (2012) A comprehensive transcriptome assembly of pigeonpea (cajanus cajan l.) using sanger and second-generation sequencing platforms. Mol Plant 5(5):1020–1028
Li H, Dong Y, Yang J (2012) De novo transcriptome of safflower and the identification of putative genes for oleosin and the biosynthesis of flavonoids. PLoS ONE 7(2):e30987
Liang C, Liu X, Yiu SM, Lim BL (2013) De novo assembly and characterization of Camelina sativa transcriptome by paired end sequencing. BMC Genomics 14:146
Lister R, Gregory BD, Ecker JR (2009) Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol 12(2):107–118
Mizrachi E, Hefer CA, Ranik M, Joubert F, Myburg AA (2010) De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genomics 11:681
Moe KT, Chung JW, Cho YI et al (2011) Sequence information on simple sequence repeats and single nucleotide polymorphisms through transcriptome analysis of Mungbean. J Integr Plant Biol 53(1):63–73
Morozova O, Hirst M, Marra MA (2009) Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet 10:135–151
Moser BR (2010) Camelina (Camelina sativa L.) oil as a biofuels feedstock: Golden opportunity or false hope? Lipid Technol 22(12):270–273
Natale DA, Shankavaram UT, Galperin MY, Wolf YI, Aravind L et al (2000) Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol 1: research0009
Natarajan P, Parani M (2011) De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GS FLX titanium platform of 454 pyrosequencing. BMC genomics 12:191
Nguyen HT, Silva JE, Podicheti R et al (2013) Camelina seed transcriptome: a tool for meal and oil improvement and translational research. Plant Biotech J 11(6):759–769
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7(2):e30619
Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38(Database issue): D822–827
Riano-Pachon DM, Correa LGG, Trejos-Espinosa R, Mueller-Roeber B (2008) Green transcription factors: a Chlamydomonas overview. Genetics 179(1):31–39
Sato S, Hirakawa H, Isobe S et al (2011) Sequence analysis of the genome of an oil-bearing tree. Jatropha curcas L. DNA Res 18(1):65–76
Schulz MH, Zerbino DR, Vingron MA, Birney EC (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28(8):1086–1092
Soriano NU Jr, Narani A (2012) Evaluation of biodiesel derived from Camelina sativa oil. J Am Oil Chem Soc 89:917–923
Tatusov RL, Natale DA, Garkavtsev IV et al (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29(1):22–28
Thudi M, Li Y, Jackson SA, May GD, Varshney RK (2012) Current state-of-art of sequencing technologies for plant genomic research. Brief Funct Genomics 11(1):3–11
Troncoso-Ponce MA, Kilaru A, Cao X et al (2011) Comparative deep transcriptional profiling of four developing oilseeds. Plant J 68(6):1014–1027
Venglat P, Xiang D, Qiu S et al (2011) Gene expression analysis of flax seed development. BMC Plant Biol 11:74
Vera JC, Wheat CW, Fescemyer HW et al (2008) Rapid transcriptome characterization for a non-model organism using 454 pyrosequencing. Mol Ecol 17(7):1636–1647
Vinogradov AE (2003) DNA helix:the importance of being GC rich. Nucleic Acids Res 31(7):1838–1844
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63
Wang Z, Fang B, Chen J et al (2010) De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genomics 11:726
Xia Z, Xu H, Zhai J et al (2011) RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol Biol 77(3):299–308
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829
Zhang J, Liang S, Duan J et al (2012) De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in Peanut (Arachis hypogaea L.). BMC Genomics 13(1):90
Zubr J, Matthaus B (2002) Effects of growth conditions on fatty acids and tocopherols in Camelina sativa oil. Ind Crops Prod 15(2):155–162
Acknowledgments
The work was funded by DST grant number (DST/IS-STAC/CO2-SR-68/09) from Department of Science and Technology, Government of India. Thanks are due to Genotypic technology (P) Ltd., Bangalore, India, for library construction, sequencing and assembly. Shalini Mudalkar is thankful to UGC, New Delhi, India, for the fellowship. Ramesh Golla was supported by Dr. D.S. Kothari Postdoctoral fellowship from UGC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shalini Mudalkar and Ramesh Golla contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mudalkar, S., Golla, R., Ghatty, S. et al. De novo transcriptome analysis of an imminent biofuel crop, Camelina sativa L. using Illumina GAIIX sequencing platform and identification of SSR markers. Plant Mol Biol 84, 159–171 (2014). https://doi.org/10.1007/s11103-013-0125-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0125-1