Abstract
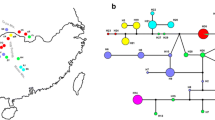
Understanding the genetic diversity and phylogenetic relationship of citron (Citrus medica L.) is of great importance for making conservation and utilization strategies. Southeast Asia and its vicinity are usually regarded as one of the centers of origin for Citrus. The principal aims of this work were to evaluate the genetic diversity of 56 accessions of citron and its relatives mainly from southwest China, to investigate the genetic structures of citron populations, and to construct a phylogenetic tree to understand the evolution of some hybrids among citrons and other citrus types. SSR analysis detected a total of 387 alleles ranging from 2 to 12 alleles per locus, and nearly all accessions identified could be unequivocally distinguished. The observed and expected heterozygosities averaged 0.36 and 0.49, respectively. From a Bayesian cluster analysis, citrons were characterized by two distinguished genetic structures, which corresponded to the geographical distribution in southwest China. Citron and fingered citron might derive from a common ancestor. Based on the chloroplast sequences, the phylogenetic trees were constructed with congruent topologies and similar levels of statistical support for relationships among citron and its relatives using both maximum parsimony and Bayesian inference methods. Citron formed a monophyletic clade, which was completely different from mandarin (C. reticulata) and pummelo (C. grandis). The genetic contributors of some hybrid species, such as C. limonia, C. aurantifolia, and C. limon, were also discussed, and citron appeared to be a primary contributor to the speciation of these secondary citrus species.





Similar content being viewed by others
References
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Barrett H, Rhodes A (1976) A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst Bot 1:105–136
Bausher MG, Singh ND, Lee S-B, Jansen RK, Daniell H (2006) The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol 6:21
Bayer RJ, Mabberley DJ, Morton C, Miller CH, Sharma IK, Pfeil BE, Rich S, Hitchcock R, Sykes S (2009) A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. Am J Bot 96:668–685
Biswas MK, Chai LJ, Mayer C, Xu Q, Guo WW, Deng XX (2012) Exploiting BAC-end sequences for the mining, characterization and utility of new short sequences repeat (SSR) markers in citrus. Mol Biol Rep 39:5373–5386
Chai LJ, Biswas MK, Yi HL, Guo WW, Deng XX (2013) Transferability, polymorphism and effectiveness for genetic mapping of the pummelo (Citrus grandis Osbeck) EST-SSR markers. Sci Hortic 155:85–91
Chen CX, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Chen HM, Jiang D, Hu ZR, Li KM, He YR, Chen SC, Li LH, Zheng DS, Liu X (2012) A newly discovered wild Ichang papeda population in Yuanjiang county, Yunnan province. J Plant Genet Res 13:929–935 (in Chinese with Engnish abstract)
Cheng YJ, Guo WW, Yi HL, Pang XM, Deng XX (2003) An efficient protocol for genomic DNA extraction from citrus species. Plant Mol Biol Report 21:177–178
de Araujo EF, de Queiroz LP, Machado MA (2003) What is citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae). Org Divers Evol 3:55–62
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Federici C, Fang D, Scora R, Roose M (1998) Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theor Appl Genet 96:812–822
Froelicher Y, Dambier D, Bassene JB, Costantino G, Lotfy S, Didout C, Beaumont V, Brottier P, Risterucci AM, Luro F, Ollitrault P (2008) Characterization of microsatellite markers in mandarin orange (Citrus reticulata Blanco). Mol Ecol Resour 8:119–122
Garcia-Lor A, Luro F, Navarro L, Ollitrault P (2012) Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Mol Gen Genet 287:77–94
Garcia-Lor A, Curk F, Snoussi-Trifa H, Morillon R, Ancillo G, Luro F, Navarro L, Ollitrault P (2013) A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Ann Bot 111:1–19
Gmitter FG, Hu X (1990) The possible role of Yunnan, China, in the origin of contemporary Citrus species (Rutaceae). Econ Bot 44:267–277
Gulsen O, Roose M (2001) Lemons: diversity and relationships with selected Citrus genotypes as measured with nuclear genome markers. J Am Soc Hortic Sci 126:309–317
Hampl V, Pavlícek A, Flegr J (2001) Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Microbiol 51:731–735
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kijas J, Thomas M, Fowler J, Roose M (1997) Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor Appl Genet 94:701–706
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948
Li XM, Xie RJ, Lu ZH, Zhou ZQ (2010) The origin of cultivated citrus as inferred from internal transcribed spacer and chloroplast DNA sequence and amplified fragment length polymorphism fingerprints. J Am Soc Hortic Sci 135:341–350
Li H, Yang XM, Zhu LS, Yi HL, Chai LJ, Deng XX (2015) Parentage analysis of natural citrus hybrid ‘Zhelong Zhoupigan’ based on nuclear and chloroplast SSR markers. Sci Hortic 186:24–30
Liang M, Yang XM, Li H, Su SY, Yi HL, Chai LJ, Deng XX (2015) De novo transcriptome assembly of pummelo and molecular marker development. PLoS One 10(3):e0120615
Lim TK (2012) Citrus medica. Edible medicinal and non-medicinal plants. Springer, Netherlands, pp 682–689
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu CH, Jiang D, Cheng YJ, Deng XX, Chen F, Fang L, Ma ZC, Xu J (2013) Chemotaxonomic study of Citrus, Poncirus and Fortunella genotypes based on peel oil volatile compounds-deciphering the genetic origin of Mangshanyegan (Citrus nobilis Lauriro). PLoS One 8(3):e58411
Long CL, Li H, Ouyang ZQ, Yang XY, Li Q, Trangmar B (2003) Strategies for agrobiodiversity conservation and promotion: a case from Yunnan, China. Biodivers Conserv 12:1145–1156
Luro FL, Costantino G, Terol J, Argout X, Allario T, Wincker P, Talon M, Ollitrault P, Morillon R (2008) Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9:287
Luro F, Venturini N, Costantino G, Paolini J, Ollitrault P, Costa J (2012) Genetic and chemical diversity of citron (Citrus medica L.) based on nuclear and cytoplasmic markers and leaf essential oil composition. Phytochemistry 77:186–196
Morton CM (2009) Phylogenetic relationships of the Aurantioideae (Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions, atpB-rbcL spacer, rps16, and trnL-trnF. Org Divers Evol 9:52–68
Morton CM, Grant M, Blackmore S (2003) Phylogenetic relationships of the Aurantioideae inferred from chloroplast DNA sequence data. Am J Bot 90:1463–1469
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 76:5269–5273
Nicolosi E, Deng Z, Gentile A, La Malfa S, Continella G, Tribulato E (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166
Nicolosi E, La Malfa S, El-Otmani M, Negbi M, Goldschmidt EE (2005) The search for the authentic citron (Citrus medica L.): historic and genetic analysis. HortSci 40:1963–1968
Ollitrault F, Terol J, Pina JA, Navarro L, Talon M, Ollitrault P (2010) Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. Am J Bot 97:124–129
Ollitrault P, Terol J, Chen C, Federici CT, Lotfy S, Hippolyte I, Ollitrault F, Berard A, Chauveau A, Cuenca J, Costantino G, Kacar Y, Mu L, Garcia-Lor A, Froelicher Y, Aleza P, Boland A, Billot C, Navarro L, Luro F, Roose ML, Gmitter FG, Talon M, Brunel D (2012a) A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics 13:593
Ollitrault P, Terol J, Garcia-Lor A, Bérard A, Chauveau A, Froelicher Y, Belzile C, Morillon R, Navarro L, Brunel D (2012b) SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics 13:13
Page R (2001) Treeview (Win32) version 1.6.6. Available at. http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
Pang XM, Hu CG, Deng XX (2006) Phylogenetic relationships within Citrus and its related genera as inferred from AFLP markers. Genet Resour Crop Evol 54:429–436
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Penjor T, Anai T, Nagano Y, Matsumoto R, Yamamoto M (2010) Phylogenetic relationships of Citrus and its relatives based on rbcL gene sequences. Tree Genet Genomes 6:931–939
Penjor T, Yamamoto M, Uehara M, Ide M, Matsumoto N, Matsumoto R, Nagano Y (2013) Phylogenetic relationships of Citrus and its relatives based on matK gene sequences. PLoS One 8(4):e62574
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quintana J, Contreras A, Merino I, Vinuesa A, Orozco G, Ovalle F, Gomez L (2015) Genetic characterization of chestnut (Castanea sativa Mill.) orchards and traditional nut varieties in El Bierzo, a glacial refuge and major cultivation site in northwestern Spain. Tree Genet Genomes 11:1–12
Ramadugu C, Keremane ML, Hu X, Karp D, Federici CT, Kahn T, Roose ML, Lee RF (2015) Genetic analysis of citron (Citrus medica L.) using simple sequence repeats and single nucleotide polymorphisms. Sci Hortic 195:124–137
Rambaut A, Drummond AJ (2007) Tracer version 1.4. Available at: http://beast.bio.ed.ac.uk/Tracer
Rana JC, Chahota RK, Sharma V, Rana M, Verma N, Verma B, Sharma TR (2015) Genetic diversity and structure of Pyrus accessions of Indian Himalayan region based on morphological and SSR markers. Tree Genet Genomes 11:1–14
Reuther W, Batchelor L, Webber H (1968) The citrus industry. University of California, USA
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system version 2.1. Exeter Software, Setauket
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruiz C, Breto MP, Asins M (2000) A quick methodology to identify sexual seedlings in citrus breeding programs using SSR markers. Euphytica 112:89–94
Scora RW (1975) On the history and origin of Citrus. Bull Torr Bot Club 102:369–375
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Sitther V, Zhang D, Harris DL, Yadav AK, Zee FT, Meinhardt LW, Dhekney SA (2014) Genetic characterization of guava (Psidium guajava L.) germplasm in the United States using microsatellite markers. Genet Resour Crop Evol 61:829–839
Snoussi H, Duval MF, Garcia-Lor A, Belfalah Z, Froelicher Y, Risterucci AM, Perrier X, Jacquemoud-Collet JP, Navarro L, Harrabi M, Ollitrault P (2012) Assessment of the genetic diversity of the Tunisian citrus rootstock germplasm. BMC Genet 13:16
Swingle WT, Reece PC (1967) The botany of Citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD (eds) The Citrus industry, vol 1. University of California, Berkeley, pp 190–430
Swofford D (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland
Tanaka T (1961) Citologia: semi-centennial commemoration papers on citrus studies. Citrologia Supporting Foundation, Osaka, p 114
Uzun A, Yesiloglu T, Polat I, Aka-Kacar Y, Gulsen O, Yildirim B, Tuzcu O, Tepe S, Canan I, Anil S (2010) Evaluation of genetic diversity in lemons and some of their relatives based on SRAP and SSR markers. Plant Mol Biol Report 29:693–701
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A 84:9054–9058
Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, Takita MA, Labadie K, Poulain J, Couloux A, Jabbari K, Cattonaro F, Del Fabbro C, Pinosio S, Zuccolo A, Chapman J, Grimwood J, Tadeo FR, Estornell LH, Munoz-Sanz JV, Ibanez V, Herrero-Ortega A, Aleza P, Perez-Perez J, Ramon D, Brunel D, Luro F, Chen C, Farmerie WG, Desany B, Kodira C, Mohiuddin M, Harkins T, Fredrikson K, Burns P, Lomsadze A, Borodovsky M, Reforgiato G, Freitas-Astua J, Quetier F, Navarro L, Roose M, Wincker P, Schmutz J, Morgante M, Machado MA, Talon M, Jaillon O, Ollitrault P, Gmitter F, Rokhsar D (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 32:656–662
Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, Chen J, Gao S, Xing F, Lan H, Chang JW, Ge X, Lei Y, Hu Q, Miao Y, Wang L, Xiao S, Biswas MK, Zeng W, Guo F, Cao H, Yang X, Xu XW, Cheng YJ, Xu J, Liu JH, Luo OJ, Tang Z, Guo WW, Kuang H, Zhang HY, Roose ML, Nagarajan N, Deng XX, Ruan Y (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45:59–66
Yeh FC, Yang R-C, Boyle TB, Ye Z, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology centre, University of Alberta, Canada
Acknowledgments
This work was funded by the National Natural Science Foundation of China (31330066 and 31221062), the Special Fund for Agro-scientific Research in the Public Interest (201303093), and the Ministry of Agriculture (CARS-27). The authors are grateful to Dr. Xiuli Zeng for collecting plant materials.
Data archiving statement
We follow standard Tree Genetics and Genomes policy. All the chloroplast gene sequences will be submitted to GenBank, and accession numbers will be provided once available.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by W.-W. Guo
This article is part of the Topical Collection on Germplasm Diversity
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Plot of Delta K (filled circles, solid line) calculated as the mean of the second-order rate of change in likelihood of K divided by the standard deviation of the likelihood of K, m(|L"(K)|)/s [L(K)] (GIF 73 kb)
Supplementary Fig. 2
Inferred clusters based on Bayesian analysis for K = 2 among 36 citron and fingered citron accessions. Each vertical line represents an individual genotype. Different colors stand for the most likely ancestry of the cluster. Individuals with multiple colors have admixed genotypes. The height of each bar represents the probability of a variety belonging to a different subgroup. The identified abbreviated accession names of corresponding samples are listed in Supplementary Table S1 (GIF 28 kb)
Supplementary Fig. 3
The 50 % majority rule consensus tree of 1502 trees obtained from four rounds of Bayesian analysis of the combined data set (matk, trnS-trnG, rps16, rpl16, atpB-rbcL, and accD-psaI chloroplast DNA regions), implementing the GTR model from 39 Citrus taxa. Branch lengths reflect changes per site. Posterior probabilities are given above branches. Clades defined as I, II, III, and IV represent different groups (GIF 54 kb)
Supplementary Table S1
Classification, origin, group, and list of studied accessions (DOCX 27 kb)
Supplementary Table S2
Summary information of the 77 nSSR primer pairs and the value of annealing temperature (T) (DOCX 20 kb)
Supplementary Table S3
The six chloroplast DNA regions used in the phylogenetic analysis, the marker names and sequences used to amplify them, the type of DNA each region contains and their annealing temperature (T) (DOCX 17 kb)
Supplementary Table S4
Heterozygosity observed in all the analyzed accessions by the 77 nSSR markers (DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Li, H., Liang, M. et al. Genetic diversity and phylogenetic relationships of citron (Citrus medica L.) and its relatives in southwest China. Tree Genetics & Genomes 11, 129 (2015). https://doi.org/10.1007/s11295-015-0955-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0955-x