Abstract
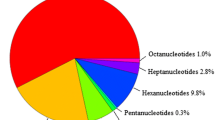
The aim of this study was to develop a large set of microsatellite markers based on publicly available BAC-end sequences (BESs), and to evaluate their transferability, discriminating capacity of genotypes and mapping ability in Citrus. A set of 1,281 simple sequence repeat (SSR) markers were developed from the 46,339 Citrus clementina BAC-end sequences (BES), of them 20.67% contained SSR longer than 20 bp, corresponding to roughly one perfect SSR per 2.04 kb. The most abundant motifs were di-nucleotide (16.82%) repeats. Among all repeat motifs (TA/AT)n is the most abundant (8.38%), followed by (AG/CT)n (4.51%). Most of the BES-SSR are located in the non-coding region, but 1.3% of BES-SSRs were found to be associated with transposable element (TE). A total of 400 novel SSR primer pairs were synthesized and their transferability and polymorphism tested on a set of 16 Citrus and Citrus relative’s species. Among these 333 (83.25%) were successfully amplified and 260 (65.00%) showed cross-species transferability with Poncirus trifoliata and Fortunella sp. These cross-species transferable markers could be useful for cultivar identification, for genomic study of Citrus, Poncirus and Fortunella sp. Utility of the developed SSR marker was demonstrated by identifying a set of 118 markers each for construction of linkage map of Citrus reticulata and Poncirus trifoliata. Genetic diversity and phylogenetic relationship among 40 Citrus and its related species were conducted with the aid of 25 randomly selected SSR primer pairs and results revealed that citrus genomic SSRs are superior to genic SSR for genetic diversity and germplasm characterization of Citrus spp.




Similar content being viewed by others
Abbreviations
- BES:
-
Citrus clementina BAC end sequences
- GSSs:
-
Genome survey sequences
- GO:
-
Gene ontology
- SSR:
-
Simple sequences repeat
References
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12:4127–4138
Schlötterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nucleic Acids Res 20:211–215
Katti MV, Ranjekar PK, Gupta VS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18:1161–1167
Mayer C, Leese F, Tollrian R (2010) Genome-wide analysis of tandem repeats in Daphnia pulex—a comparative approach. BMC Genomics 11:277
Shanker A, Bhargava A, Bajpai R, Singh S, Srivastava S, Sharma V (2007) Bioinformatically mined simple sequence repeats in UniGene of Citrus sinensis. Sci Hortic 113:353–361
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Chen C, Yu Q, Hou S, Li Y, Eustice M, Skelton RL, Veatch O, Herdes RE, Diebold L, Saw J, Feng Y, Qian W, Bynum L, Wang L, Moore PH, Paull RE, Alam M, Ming R (2007) Construction of a sequence-tagged high-density genetic map of papaya for comparative structural and evolutionary genomics in brassicales. Genetics 177:2481–2491
Shoemaker RC, Grant D, Olson T, Warren WC, Wing R, Yu Y, Kim H, Cregan P, Joseph B, Futrell-Griggs M, Nelson W, Davito J, Walker J, Wallis J, Kremitski C, Scheer D, Clifton SW, Graves T, Nguyen H, Wu X, Luo M, Dvorak J, Nelson R, Cannon S, Tomkins J, Schmutz J, Stacey G, Jackson S (2008) Microsatellite discovery from BAC end sequences and genetic mapping to anchor the soybean physical and genetic maps. Genome 51:294–302
Shultz JL, Kazi S, Bashir R, Afzal JA, Lightfoot DA (2007) The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theor Appl Genet 114:1081–1090
Pan L, Xia Q, Quan Z, Liu H, Ke W, Ding Y (2010) Development of novel EST-SSRs from Sacred Lotus (Nelumbo nucifera Gaertn) and their utilization for the genetic diversity analysis of N. nucifera. J Hered 101:71–82
Batley J, Hopkins CJ, Cogan NOI, Hand M, Jewell E, Kaur J, Kaur S, Li XI, Ling AE, Love C, Mountford H, Todorovic M, Vardy M, Walkiewicz M, Spangenberg GC, Edwards D (2007) Identification and characterization of simple sequence repeat markers from Brassica napus expressed sequences. Mol Ecol Notes 7:886–889
Choi SR, Teakle GR, Plaha P, Kim JH, Allender CJ, Beynon E, Piao ZY, Soengas P, Han TH, King GJ, Barker GC, Hand P, Lydiate DJ, Batley J, Edwards D, Koo DH, Bang JW, Park BS, Lim YP (2007) The reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Theor Appl Genet 115:777–792
Hopkins CJ, Cogan NOI, Hand M, Jewell E, Kaur J, Li XI, Lim GAC, Ling AE, Love C, Mountford H, Todorovic M, Vardy M, Spangenberg GC, Edwards D, Batley J (2007) Sixteen new simple sequence repeat markers from Brassica juncea expressed sequences and their cross-species amplification. Mol Ecol Notes 7:697–700
Iniguez-Luy FL, Voort AV, Osborn TC (2008) Development of a set of public SSR markers derived from genomic sequence of a rapid cycling Brassica oleracea L. genotype. Theor Appl Genet 117:977–985
Ling AE, Kaur J, Burgess B, Hand M, Hopkins CJ, Li XI, Love CG, Vardy M, Walkiewicz M, Spangenberg G, Edwards D, Batley J (2007) Characterization of simple sequence repeat markers derived in silico from Brassica rapa bacterial artificial chromosome sequences and their application in Brassica napus. Mol Ecol Notes 7:273–277
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104:399–407
Peng J, Lapitan N (2005) Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct Integr Genomics 5:80–96
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Hackauf B, Wehling P (2002) Identification of microsatellite polymorphisms in an expressed portion of the rye genome. Plant Breed 121:17–25
Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, Gardiner J, Bergstrom D, Houchins K, Melia-Hancock S, Musket T, Duru N, Polacco M, Edwards K, Ruff T, Register JC, Brouwer C, Thompson R, Velasco R, Chin E, Lee M, Woodman-Clikeman W, Long MJ, Liscum E, Cone K, Davis G, Coe EH Jr (2002) Development and mapping of SSR markers for maize. Plant Mol Biol 48:463–481
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160:1115–1123
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. TAG Theor Appl Genet 100:723–726
Castelo AT, Martins W, Gao GR (2002) TROLL—tandem repeat occurrence locator. Bioinformatics 18:634–636
Luro F, Costantino G, Terol J, Argout X, Allario T, Wincker P, Talon M, Ollitrault P, Morillon R (2008) Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9:287
Kijas JMH, Thomas MR, Fowler JCS, Roose ML (1997) Integration of trinucleotide microsatellites into a linkage map of Citrus. TAG Theor Appl Genet 94:701–706
Jiang D, Zhong G-Y, Hong Q-B (2006) Analysis of microsatellites in citrus unigenes. Acta Genetica Sinica 33:345–353
Novelli VM, Cristofani M, Souza AA, Machado MA (2006) Development and characterization of polymorphic microsatellite markers for the sweet orange (Citrus sinensis L. Osbeck). Genet Mol Biol 29:90–96
Terol J, Naranjo MA, Ollitrault P, Talon M (2008) Development of genomic resources for Citrus clementina: characterization of three deep-coverage BAC libraries and analysis of 46, 000 BAC end sequences. BMC Genomics 9:423
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Ollitrault F, Terol J, Pina JA, Navarro L, Talon M, Ollitrault P (2010) Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. Am J Bot 97:124–129
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Cheng Y-J, Guo W–W, Yi H-L, Pang X-M, Deng X (2003) An efficient protocol for genomic DNA extraction from Citrus species. Plant Mol Biol Rep 21:177–178
Ruiz C, Paz Breto M, Asíns MJ (2000) A quick methodology to identify sexual seedlings in citrus breeding programs using SSR markers. Euphytica 112:89–94
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Labarga A, Valentin F, Anderson M, Lopez R (2007) Web services at the European bioinformatics institute. Nucleic Acids Res 35:W6–W11
Rohlf FJ (1998) NTSYSpc: numerical taxonomy and multivariate analysis system version 2.02. Exeter Software, Setauket
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Corazza-Nunes MJ, Machado MA, Nunes WMC, Cristofani M, Targon MLPN (2002) Assessment of genetic variability in grapefruits (Citrus paradisi Macf.) and pummelos (C. maxima (Burm.) Merr.) using RAPD and SSR markers. Euphytica 126:169–176
Hong CP, Piao ZY, Kang TW, Batley J, Yang TJ, Hur YK, Bhak J, Park BS, Edwards D, Lim YP (2007) Genomic distribution of simple sequence repeats in Brassica rapa. Mol Cells 23:349–356
Wen M, Wang H, Xia Z, Zou M, Lu C, Wang W (2010) Developmenrt of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha Curcas L. BMC Res Notes 3:42
Leclercq S, Rivals E, Jarne P (2007) Detecting microsatellites within genomes: significant variation among algorithms. BMC Bioinform 8:125
Lai C, Yu Q, Hou S, Skelton R, Jones M, Lewis K, Murray J, Eustice M, Guan P, Agbayani R, Moore P, Ming R, Presting G (2006) Analysis of papaya BAC end sequences reveals first insights into the organization of a fruit tree genome. Mol Genet Genomics 276:1–12
Gao L, Tang J, Li H, Jia J (2003) Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed 12:245–261
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Tóth G, Gáspári Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Labbé J, Murat C, Morin E, Le Tacon F, Martin F (2011) Survey and analysis of simple sequence repeats in the Laccaria bicolor genome, with development of microsatellite markers. Curr Genet 57:75–88
Victoria F, da Maia L, de Oliveira A (2011) In silico comparative analysis of SSR markers in plants. BMC Plant Biol 11:15
Akagi H, Yokozeki Y, Inagaki A, Mori K, Fujimura T (2001) Micron a microsatellite-targeting transposable element in the rice genome. Mol Genet Genomics 266:471–480
Ramsay L, Macaulay M, Cardle L, Morgante M, Ivanissevich SD, Maestri E, Powell W, Waugh R (1999) Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J 17:415–425
Tay W, Behere G, Batterham P, Heckel D (2010) Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evol Biol 10:144
Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I (2003) Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet 107:736–744
Barkley NA, Roose ML, Krueger RR, Federici CT (2006) Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor Appl Genet 112:1519–1531
Yong L, De-Chun L, Bo W, Zhong-Hai S (2006) Genetic diversity of pummelo (Citrus grandis Osbeck) and its relatives based on simple sequence repeat markers. Chin J Agric Biotechnol 3:119–126
Zhang TP, Peng SL, Wang ZF, Ling DH, Gan LS (2001) Genetic relationships among cultivars of citrus maxima (burm.) Merr. using RAPD marker technique. J Trop Sub Trop Bot 9:322–328
Bernet GP, Margaix C, Jacas J, Carbonell EA, Asins MJ (2005) Genetic analysis of citrus leafminer susceptibility. Theor Appl Genet 110:1393–1400
Ruiz C, Asins MJ (2003) Comparison between Poncirus and Citrus genetic linkage maps. Theor Appl Genet 106:826–836
Siviero A, Cristofani M, Furtado EL, Garcia AA, Coelho AS, Machado MA (2006) Identification of QTLs associated with citrus resistance to Phytophthora gummosis. J Appl Genet 47:23–28
Weber CA, Moore GA, Deng Z, Gmitter FG Jr (2003) Mapping freeze tolerance quantitative trait loci in a Citrus grandis × Poncirus trifoliata F1 pseudo-testcross using molecular markers. J Am Soc Hortic Sci 128:508–514
Cai Q, Guy CL, Moore GA (1994) Extension of the linkage map in Citrus using random amplified polymorphic DNA (RAPD) markers and RFLP mapping of cold-acclimation-responsive loci. TAG Theor Appl Genet 89:606–614
Durham RE, Liou PC, Gmitter FG, Moore GA (1992) Linkage of restriction fragment length polymorphisms and isozymes in Citrus. TAG Theor Appl Genet 84:39–48
Jarrell DC, Roose ML, Traugh SN, Kupper RS (1992) A genetic map of citrus based on the segregation of isozymes and RFLPs in an intergeneric cross. TAG Theor Appl Genet 84:49–56
Moore GA, Tozlu I, Weber CA, Guy CL (2000) Mapping quantitative trait loci for salt tolerance and cold tolerance in Citrus grandis (L.) Osb. × Poncirus trifoliata (L.) Raf hybrid populations. Acta Hortic 535:37–45
Fraser LG, Harvey CF, Crowhurst RN, De Silva HN (2004) EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016
Pang XM, Hu CG, Deng XX (2003) Phylogenetic relationships among citrus and its relatives as reveled by SSR markers. Yi Chuan Bao 30(1):81–87
Acknowledgments
We are grateful to Dr. Yun-Jiang Cheng for discussing on the sampling strategy. This research was financially supported by the Ministry of Science and Technology of China (Nos. 2011CB100600, 2011AA100205) and the national NSF of China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Biswas, M.K., Chai, L., Mayer, C. et al. Exploiting BAC-end sequences for the mining, characterization and utility of new short sequences repeat (SSR) markers in Citrus. Mol Biol Rep 39, 5373–5386 (2012). https://doi.org/10.1007/s11033-011-1338-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1338-5