Abstract
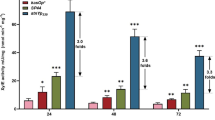
The selection of efficient promoter is usually very crucial for gene expression and metabolic engineering in Streptomycetes. In this study, the synthetic promoters SPL-57and SPL-21, and the engineered promoter kasOp*were selected and their activities were examined by using a reporter gene assay based on GUS. All selected promoters which have been reported to be stronger than promoter permE*, which was used as control promoter. As host we were choosing S. diastatochromogenes 1628, the producer of toyocamycin (TM). Our results indicate that all tested promoters can be used to express genes in S. diastatochromogenes 1628. Interesting, promoter SPL-21 showed the strongest transcriptional and expression level and gave rise to a 5.2-fold increase in GUS activity compared with control. In order to improve TM production, the promoters were used to control expression of toyF. This gene encodes an adenylosuccinate lyase involved in TM biosynthesis. Among all different recombinant strains, the strain 1628-21F, in which over-expression of toyF gene was driven by SPL-21, exhibited the largest increase in TOYF activity and TM production. In a 5-l fermenter this strain produced more than two times more TM compared with the wild-type strain.




Similar content being viewed by others
References
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Battaglia U, Long JE, Searle MS, Moody CJ (2011) 7-Deazapurine biosynthesis: NMR study of toyocamycin biosynthesis in Streptomyces rimosus using 2-13C-7-15N-adenine. Org Biomol Chem 9:2227–2232
Braatsch S, Helmark S, Kranz H, Koebmann B, Jensen PR (2008) Escherichia coli strains with promoter libraries constructed by Red/ET recombination pave the way for transcriptional fine-tuning. Biotechniques 45:335–337
Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H (2010) The complex extracellular biology of Streptomyces. FEMS Microbiol Rev 34:171–198
Chiang YM, Chang SL, Oakley BR, Wang CC (2011) Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol 15:137–143
Gomez-Escribano JP, Bibb MJ (2014) Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol 41:425–431
Hammer K, Mijakovic I, Jensen PR (2006) Synthetic promoter libraries–tuning of gene expression. Trends Biotechnol 24:53–55
Kanth BK, Jnawali HN, Niraula NP, Sohng JK (2011) Superoxide dismutase (SOD) genes in Streptomyces peucetius: effects of SODs on secondary metabolites production. Microbiol Res 166:391–402
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical streptomyces genetics. John Innes Foundation, Norwich
Lanzer M, Bujard H (1988) Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA 85:8973–8977
Li J, Zhang Y (2014) Relationship between promoter sequence and its strength in gene expression. Eur Phys J E Soft Matter 37:44
Liu H, Bao X (2009) Overexpression of the chitosanase gene in Fusarium solani via Agrobacterium tumefaciens-mediated transformation. Curr Microbiol 58:279–282
Ma Z, Liu J, Bechthold A, Tao L, Shentu X, Bian Y, Yu X (2014a) Development of intergeneric conjugal gene transfer system in Streptomyces diastatochromogenes 1628 and its application for improvement of toyocamycin production. Curr Microbiol 68:180–185
Ma Z, Tao L, Bechthold A, Shentu X, Bian Y, Yu X (2014b) Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl Microbiol Biotechnol 98:5051–5058
Ma Z, Luo S, Xu X, Bechthold A, Yu X (2016) Characterization of representative rpoB gene mutations leading to a signifcant change in toyocamycin production of Streptomyces diastatochromogenes 1628. J Ind Microbiol Biotechnol 43:463–471
Martin JF, Liras P (2010) Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 13:263–273
Mccarty RM, Bandarian V (2008) Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem Biol 15:790–798
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) Beta-glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77:5370–5383
Niraula NP, Shrestha P, Oh TJ, Sohng JK (2010) Identification and characterization of a NADH oxidoreductase involved in phenylacetic acid degradation pathway from Streptomyces peucetius. Microbiol Res 165:649–656
Rodriguez H, Rico S, Diaz M, Santamaria RI (2013) Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microb Cell Fact 12:127
Seghezzi N, Amar P, Koebmann B, Jensen PR, Virolle MJ (2011) The construction of a library of synthetic promoters revealed some specific features of strong Streptomyces promoters. Appl Microbiol Biotechnol 90:615–623
Shrestha P, Oh TJ, Liou K, Sohng JK (2008) Cytochrome P450 (CYP105F2) from Streptomyces peucetius and its activity with oleandomycin. Appl Microbiol Biotechnol 79:555–562
Siegl T, Tokovenko B, Myronovskyi M, Luzhetskyy A (2013) Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab Eng 19:98–106
Sohoni SV, Fazio A, Workman CT, Mijakovic I, Lantz AE (2014) Synthetic promoter library for modulation of actinorhodin production in Streptomyces coelicolor A3(2). PLoS ONE 9:e99701
Strohl WR (1992) Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res 20:961–974
Tao L, Ma Z, Xu X, Andreas B, Bian Y, Shentu X, Yu X (2015) Engineering Streptomyces diastatochromogenes 1628 to increase the production of toyocamycin. Eng. Life Sci 15:779–787
Wang T, Bai L, Zhu D, Lei X, Liu G, Deng Z, You D (2012) Enhancing macrolide production in Streptomyces by coexpressing three heterologous genes. Enzyme Microb Technol 50:5–9
Wang W, Li X, Wang J, Xiang S, Feng X, Yang K (2013) An engineered strong promoter for streptomycetes. Appl Environ Microbiol 79:4484–4492
Woodward DO (1978) Adenylosuccinate AMP-lyase (Neurospora crassa). Methods Enzymol 51:202–207
Xu XH, Tao LB, Ma Z, Yu XP (2016) Cloning of toyF gene and its relevance to toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Chin J. Biol Control 32:511–517 (Chinese)
Zhang B, Yang D, Yan Y, Pan G, Xiang W, Shen B (2016) Overproduction of lactimidomycin by cross-overexpression of genes encoding Streptomyces antibiotic regulatory proteins. Appl Microbiol Biotechnol 100:2267–2277
Zhu H, Sandiford SK, van Wezel GP (2014) Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41:371–386
Acknowledgements
This work was supported by National Natural Science Foundation of China (31401792), Fund for excellent youth of Zhejiang province, China (LR17C140002). We thank Prof. Yang K.Q. and Prof. Luztheskyy A. for providing plasmids and bacterial strain.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, X., Wang, J., Bechthold, A. et al. Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J Microbiol Biotechnol 33, 30 (2017). https://doi.org/10.1007/s11274-016-2194-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2194-1