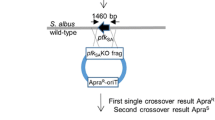
Abstract
Heterologous gene expression is one of the main strategies used to access the full biosynthetic potential of actinomycetes, as well as to study the metabolic pathways of natural product biosynthesis and to create unnatural pathways. Streptomyces coelicolor A3(2) is the most studied member of the actinomycetes, bacteria renowned for their prolific capacity to synthesize a wide range of biologically active specialized metabolites. We review here the use of strains of this species for the heterologous production of structurally diverse actinomycete natural products.
Similar content being viewed by others
References
Bérdy J (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (Tokyo) 65:385–395
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335
Baltz RH (2007) Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563
W-H Li J, Vederas JC (2009) Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165
Sources of therapeutically useful antibiotics. It is difficult to obtain accurate information about the number of marketed antibiotics obtained directly from, or derived from metabolites produced by, actinomycetes. Our figures were obtained by analysing the origin of the antibiotics listed on http://en.wikipedia.org/wiki/Timeline_of_antibiotics and classifying them into two categories, “fully synthetic” and “natural product related”; the last category was further divided according to the producing organism into three categories, “actinomycete”, “other bacteria” and “fungi”. The initial list contains 138 antibiotic formulations, of which 28 are based on a fully synthetic active pharmaceutical ingredient (API) (mostly quinolones and fluoroquinolones) and 110 contain APIs derived from natural products (80%). Of these 110, 48 formulations (44% of all natural product formulations; 35% of all formulations) contain metabolites isolated from actinomycetes (or semisynthetic derivatives of them), three are derived from other bacteria, and 59 contain APIs derived from metabolites produced by fungi (almost exclusively semisynthetic compounds derived from penicillins and cephalosporins). Therefore, 35% of all marketed antibiotic formulations contain an active ingredient derived, directly or indirectly, from an actinomycete. Our analysis suggests that all marketed APIs are derived from just 41 original molecules, 33 of which are natural products; only five of these 33 are produced by fungi and three by bacteria other than actinomycetes, leaving 25 original structures produced by actinomycetes. Thus actinomycetes appear to be the source of 61% of all original molecules and of 76% of the original natural product compounds developed for use in marketed antibiotic formulations
Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang H, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Challis GL, Hopwood DA (2003) Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA 100(Suppl 2):14555–14561
Challis GL (2008) Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 154:1555–1569
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384
Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E (2010) Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156:2343–2353
Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA 108:6258–6263
Ochi K, Tanaka Y, Tojo S (2013) Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymeraseor or by rare earth elements. J Indust Microbiol Biotechnol
Zhu H, Sandifod SK, van Wezel GP (2013) Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol
Gomez-Escribano JP, Bibb MJ (2012) Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol 517:279–300
Baltz RH (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37:759–772
Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF (2004) Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54:107–128
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA, Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (eds) (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Jones AC, Gust B, Kulik A, Heide L, Buttner MJ, Bibb MJ (2013) Phage p1-derived artificial chromosomes facilitate heterologous expression of the FK506 gene cluster. PLoS One 8:e69319
Flärdh K, Buttner MJ (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49
Barona-Gómez F, Lautru S, Francou F-X, Leblond P, Pernodet J-L, Challis GL (2006) Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 152:3355–3366
Barona-Gómez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL (2004) Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J Am Chem Soc 126:16282–16283
Bystrykh LV, Fernández-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L (1996) Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2). J Bacteriol 178:2238–2244
Gomez-Escribano JP, Song L, Fox DJ, Yeo V, Bibb MJ, Challis GL (2012) Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem Sci 3:2716–2720
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546
Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BAM, Hayes MA, Smith CP, Micklefield J (2002) Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9:1175–1187
Jiang J, He X, Cane DE (2007) Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat Chem Biol 3:711–715
Kempter C, Kaiser D, Haag S, Nicholson G, Gnau V, Walk T, Gierling KH, Decker H, Zähner H, Jung G, Metzger JW (1997) CDA: calcium-dependent peptide antibiotics from Streptomyces coelicolor A3(2) containing unusual residues. Angew Chem Int Ed 36:498–501
Lautru S, Deeth RJ, Bailey LM, Challis GL (2005) Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol 1:265–269
Lin X, Hopson R, Cane DE (2006) Genome mining in Streptomyces coelicolor: molecular cloning and characterization of a new sesquiterpene synthase. J Am Chem Soc 128:6022–6023
Mo S, Sydor PK, Corre C, Alhamadsheh MM, Stanley AE, Haynes SW, Song L, Reynolds KA, Challis GL (2008) Elucidation of the Streptomyces coelicolor pathway to 2-undecylpyrrole, a key intermediate in undecylprodiginine and streptorubin B biosynthesis. Chem Biol 15:137–148
Song L, Barona-Gomez F, Corre C, Xiang L, Udwary DW, Austin MB, Noel JP, Moore BS, Challis GL (2006) Type III polyketide synthase beta-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J Am Chem Soc 128:14754–14755
Taguchi T, Itou K, Ebizuka Y, Malpartida F, Hopwood DA, Surti CM, Booker-Milburn KI, Stephenson GR, Ichinose K (2000) Chemical characterisation of disruptants of the Streptomyces coelicolor A3(2) actVI genes involved in actinorhodin biosynthesis. J Antibiot (Tokyo) 53:144–152
Tsao SW, Rudd BA, He XG, Chang CJ, Floss HG (1985) Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J Antibiot (Tokyo) 38:128–131
Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE (2008) Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2). J Biol Chem 283:8183–8189
Flinspach K, Westrich L, Kaysser L, Siebenberg S, Gomez-Escribano JP, Bibb M, Gust B, Heide L (2010) Heterologous expression of the biosynthetic gene clusters of coumermycin A(1), clorobiocin and caprazamycins in genetically modified Streptomyces coelicolor strains. Biopolymers 93:823–832
Sohoni SV, Bapat PM, Lantz AE (2012) Robust, small-scale cultivation platform for Streptomyces coelicolor. Microb Cell Fact 11:9
McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C (1993) Engineered biosynthesis of novel polyketides. Science 262:1546–1550
Floriano B, Bibb M (1996) afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 21:385–396
Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97:87–98
Young TS, Dorrestein PC, Walsh CT (2012) Codon randomization for rapid exploration of chemical space in thiopeptide antibiotic variants. Chem Biol 19:1600–1610
Wyszynski FJ, Hesketh AR, Bibb MJ, Davis BG (2010) Dissecting tunicamycin biosynthesis by genome mining: cloning and heterologous expression of a minimal gene cluster. Chem Sci 1:581–589
Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, Bibb MJ, Lee SJ, Davies GJ, Davis BG (2012) Biosynthesis of the tunicamycin antibiotics proceeds via unique exo-glycal intermediates. Nat Chem 4:539–546
Völler GH, Krawczyk JM, Pesic A, Krawczyk B, Nachtigall J, Süssmuth RD (2012) Characterization of new class III lantibiotics–erythreapeptin, avermipeptin and griseopeptin from Saccharopolyspora erythraea, Streptomyces avermitilis and Streptomyces griseus demonstrates stepwise N-terminal leader processing. Chembiochem 13:1174–1183
Li T, Du Y, Cui Q, Zhang J, Zhu W, Hong K, Li W (2013) Cloning, characterization and heterologous expression of the indolocarbazole biosynthetic gene cluster from marine-derived Streptomyces sanyensis FMA. Mar Drugs 11:466–488
Kaysser L, Bernhardt P, Nam S-J, Loesgen S, Ruby JG, Skewes-Cox P, Jensen PR, Fenical W, Moore BS (2012) Merochlorins A-D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J Am Chem Soc 134:11988–11991
Huo L, Rachid S, Stadler M, Wenzel SC, Müller R (2012) Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis. Chem Biol 19:1278–1287
Hopwood DA, Malpartida F, Kieser HM, Ikeda H, Duncan J, Fujii I, Rudd BA, Floss HG, Omura S (1985) Production of ‘hybrid’ antibiotics by genetic engineering. Nature 314:642–644
Alt S, Burkard N, Kulik A, Grond S, Heide L (2011) An artificial pathway to 3,4-dihydroxybenzoic acid allows generation of new aminocoumarin antibiotic recognized by catechol transporters of E. coli. Chem Biol 18:304–313
Wright F, Bibb MJ (1992) Codon usage in the G+C-rich Streptomyces genome. Gene 113:55–65
Saleh O, Flinspach K, Westrich L, Kulik A, Gust B, Fiedler H-P, Heide L (2012) Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663. Beilstein J Org Chem 8:501–513
Saleh O, Bonitz T, Flinspach K, Kulik A, Burkard N, Muhlenweg A, Vente A, Polnick S, Lammerhofer M, Gust B, Fiedler H-P, Heide L (2012) Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Med Chem Commun 3:1009–1019
Smanski MJ, Casper J, Peterson RM, Yu Z, Rajski SR, Shen B (2012) Expression of the platencin biosynthetic gene cluster in heterologous hosts yielding new platencin congeners. J Nat Prod 75:2158–2167
Du D, Zhu Y, Wei J, Tian Y, Niu G, Tan H (2013) Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl Microbiol Biotechnol 97:6383–6396
Bell R (2010) Analysis and manipulation of “actagardine” gene clusters from Actinoplanes. PhD thesis, University of East Anglia
Boakes S, Cortés J, Appleyard AN, Rudd BAM, Dawson MJ (2009) Organization of the genes encoding the biosynthesis of actagardine and engineering of a variant generation system. Mol Microbiol 72:1126–1136
Foulston LC, Bibb MJ (2010) Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc Natl Acad Sci USA 107:13461–13466
Foulston L (2010) Cloning and analysis of the microbisporicin lantibiotic gene cluster from Microbispora corallina. PhD thesis University of East Anglia
Foulston L, Bibb M (2011) Feed-forward regulation of microbisporicin biosynthesis in Microbispora corallina. J Bacteriol 193:3064–3071
Sherwood EJ, Bibb MJ (2013) The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. Proc Natl Acad Sci USA 110:E2500–E2509
Sherwood EJ, Hesketh AR, Bibb MJ (2013) Cloning and analysis of the planosporicin lantibiotic biosynthetic gene cluster of Planomonospora alba. J Bacteriol 195:2309–2321
Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-Ya K, Cane DE, Ikeda H (2013) Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol 2:384–396
Marcone GL, Foulston L, Binda E, Marinelli F, Bibb M, Beltrametti F (2010) Methods for the genetic manipulation of Nonomuraea sp. ATCC 39727. J Ind Microbiol Biotechnol 37:1097–1103
Claesen J, Bibb M (2010) Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA 107:16297–16302
Claesen J, Bibb MJ (2011) Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J Bacteriol 193:2510–2516
Kaysser L, Tang X, Wemakor E, Sedding K, Hennig S, Siebenberg S, Gust B (2011) Identification of a napsamycin biosynthesis gene cluster by genome mining. Chembiochem 12:477–487
Niu G, Li L, Wei J, Tan H (2013) Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chem Biol 20:34–44
Jankowitsch F, Schwarz J, Rückert C, Gust B, Szczepanowski R, Blom J, Pelzer S, Kalinowski J, Mack M (2012) Genome sequence of the bacterium Streptomyces davawensis JCM 4913 and heterologous production of the unique antibiotic roseoflavin. J Bacteriol 194:6818–6827
Robles-Reglero V, Santamarta I, Álvarez-Álvarez R, Martín JF, Liras P (2013) Transcriptional analysis and proteomics of the holomycin gene cluster in overproducer mutants of Streptomyces clavuligerus. J Biotechnol 163:69–76
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gomez-Escribano, J.P., Bibb, M.J. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol 41, 425–431 (2014). https://doi.org/10.1007/s10295-013-1348-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1348-5