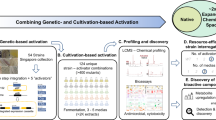
Abstract
Actinomycetes are a rich source of natural products, and these mycelial bacteria produce the majority of the known antibiotics. The increasing difficulty to find new drugs via high-throughput screening has led to a decline in antibiotic research, while infectious diseases associated with multidrug resistance are spreading rapidly. Here we review new approaches and ideas that are currently being developed to increase our chances of finding novel antimicrobials, with focus on genetic, chemical, and ecological methods to elicit the expression of biosynthetic gene clusters. The genome sequencing revolution identified numerous gene clusters for natural products in actinomycetes, associated with a potentially huge reservoir of unknown molecules, and prioritizing them is a major challenge for in silico screening-based approaches. Some antibiotics are likely only expressed under very specific conditions, such as interaction with other microbes, which explains the renewed interest in soil and marine ecology. The identification of new gene clusters, as well as chemical elicitors and culturing conditions that activate their expression, should allow scientists to reinforce their efforts to find the necessary novel antimicrobial drugs.




Similar content being viewed by others
References
Aharonowitz Y (1980) Nitrogen metabolite regulation of antibiotic biosynthesis. Annu Rev Microbiol 34:209–233
Alexander DC, Rock J, He X, Brian P, Miao V, Baltz RH (2010) Development of a genetic system for combinatorial biosynthesis of lipopeptides in Streptomyces fradiae and heterologous expression of the A54145 biosynthesis gene cluster. Appl Environ Microbiol 76:6877–6887
Allenby NE, Laing E, Bucca G, Kierzek AM, Smith CP (2012) Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic acids Res 40:9543–9556
Altermann E, Klaenhammer TR (2005) PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC genomics 6:60
Angell S, Lewis CG, Buttner MJ, Bibb MJ (1994) Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol Gen Genet 244:135–143
Angell S, Schwarz E, Bibb MJ (1992) The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol 6:2833–2844
Aparicio JF, Colina AJ, Ceballos E, Martin JF (1999) The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. A new polyketide synthase organization encoded by two subclusters separated by functionalization genes. J Biol Chem 274:10133–10139
Arias CA, Murray BE (2009) Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N Engl J Med 360:439–443
Bachmann BO, Ravel J (2009) Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol 458:181–217
Baltz RH (2007) Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131
Baltz RH (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol 38:1747–1760
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Current Opin Pharmacol 8:557–563
Beltrametti F, Rossi R, Selva E, Marinelli F (2006) Antibiotic production improvement in the rare actinomycete Planobispora rosea by selection of mutants resistant to the aminoglycosides streptomycin and gentamycin and to rifamycin. J Ind Microbiol Biotechnol 33:283–288
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bérdy J (2005) Bioactive microbial metabolites. J Antibiot (Tokyo) 58:1–26
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Bibb MJ, Hesketh A (2009) Chapter 4. Analyzing the regulation of antibiotic production in streptomycetes. Methods Enzymol 458:93–116
Birko Z, Bialek S, Buzas K, Szajli E, Traag BA, Medzihradszky KF, Rigali S, Vijgenboom E, Penyige A, Kele Z, van Wezel GP, Biro S (2007) The secreted signaling protein factor C triggers the A-factor response regulon in Streptomyces griseus: overlapping signaling routes. Mol Cell Proteomics 6:1248–1256
Biro S, Bekesi I, Vitalis S, Szabo G (1980) A substance effecting differentiation in Streptomyces griseus. Purification and properties. Eur J Biochem 103:359–363
Biro S, Birko Z, van Wezel GP (2000) Transcriptional and functional analysis of the gene for factor C, an extracellular signal protein involved in cytodifferentiation of Streptomyces griseus. Antonie Van Leeuwenhoek 78:277–285
Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP (2009) Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol 5:462–464
Brückner R, Titgemeyer F (2002) Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett 209:141–148
Caboche S, Pupin M, Leclere V, Fontaine A, Jacques P, Kucherov G (2008) NORINE: a database of nonribosomal peptides. Nucleic Acids Res 36:D326–D331
Carter RA, Worsley PS, Sawers G, Challis GL, Dilworth MJ, Carson KC, Lawrence JA, Wexler M, Johnston AW, Yeoman KH (2002) The vbs genes that direct synthesis of the siderophore vicibactin in Rhizobium leguminosarum: their expression in other genera requires ECF sigma factor RpoI. Mol Microbiol 44:1153–1166
Challis GL, Hopwood DA (2003) Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA 100:14555–14561
Charusanti P, Fong NL, Nagarajan H, Pereira AR, Li HJ, Abate EA, Su Y, Gerwick WH, Palsson BO (2012) Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. PLoS One 7:e33727
Chater KF (2006) Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philosophical Transactions of the Royal Society of London. Ser B Biol Sci 361:761–768
Chuanchuen R, Schweizer HP (2012) Global transcriptional responses to triclosan exposure in Pseudomonas aeruginosa. Int J Antimicrob Agents 40:114–122
Colson S, Stephan J, Hertrich T, Saito A, van Wezel GP, Titgemeyer F, Rigali S (2007) Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J Mol Microbiol Biotechnol 12:60–66
Colson S, van Wezel GP, Craig M, Noens EE, Nothaft H, Mommaas AM, Titgemeyer F, Joris B, Rigali S (2008) The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154:373–382
Corre C, Song L, O’Rourke S, Chater KF, Challis GL (2008) 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc Natl Acad Sci USA 105:17510–17515
Craig M, Lambert S, Jourdan S, Tenconi E, Colson S, Maciejewska M, Martin JF, Ongena M, van Wezel G, Rigali S (2012) Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Environ Microbiol Rep 4:512–521
Craney A, Ozimok C, Pimentel-Elardo SM, Capretta A, Nodwell JR (2012) Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem Biol 19:1020–1027
D’Alia D, Eggle D, Nieselt K, Hu WS, Breitling R, Takano E (2011) Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3(2). Microb Biotechnol 4:239–251
de Jong A, van Heel AJ, Kok J, Kuipers OP (2010) BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res 38:W647–W651
Demain AL (1989) Carbon source regulation of idiolite biosynthesis in regulation of secondary metabolism in actinomycetes. CRC Press, Boca Raton, pp 127–134
Demain AL (1999) Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol 52:455–463
Dobretsov S, Dahms HU, Yili H, Wahl M, Qian PY (2007) The effect of quorum-sensing blockers on the formation of marine microbial communities and larval attachment. FEMS Microbiol Ecol 60:177–188
Dulaney EL (1948) Observations on Streptomyces griseus: II. Nitrogen sources for growth and streptomycin production. J Bacteriol 56:305–313
Fedorova D, Moktali V, Medema H (2012) Bioinformatics approaches and software for detection of secondary metabolic gene clusters. In: Keller NP, Turner G (eds) Fungal secondary metabolism, vol. 944. Humana Press, New York, pp 23–45
Flärdh K, Buttner MJ (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49
Fleming A (1929) The antibacterial action of a Penicillium, with special reference to their use for the isolation of B. influenzae. Brit J Exp Pathol 10:226–236
Floriano B, Bibb M (1996) afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 21:385–396
Goosen N, van de Putte P (1995) The regulation of transcription initiation by integration host factor. Mol Microbiol 16:1–7
Görke B, Stülke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624
Gross H (2009) Genomic mining—a concept for the discovery of new bioactive natural products. Curr Opin Drug Discov Dev 12:207–219
Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP (2012) Identification of glucose kinase dependent and independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol Microbiol 86:1490–1507
Hara H, Ohnishi Y, Horinouchi S (2009) DNA microarray analysis of global gene regulation by A-factor in Streptomyces griseus. Microbiology 155:2197–2210
Hiard S, Maree R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Rigali S (2007) PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun 357:861–864
Hirano S, Tanaka K, Ohnishi Y, Horinouchi S (2008) Conditionally positive effect of the TetR-family transcriptional regulator AtrA on streptomycin production by Streptomyces griseus. Microbiology 154:905–914
Hopwood DA (1999) Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145:2183–2202
Hopwood DA (2006) Soil to genomics: the Streptomyces chromosome. Annu Rev Genet 40:1–23
Hopwood DA (2007) Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York
Horinouchi S, Beppu T (1992) Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol 46:377–398
Horinouchi S, Beppu T (1994) A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol 12:859–864
Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K (2009) Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27:462–464
Hsiao NH, Gottelt M, Takano E (2009) Chapter 6. Regulation of antibiotic production by bacterial hormones. Methods Enzymol 458:143–157
Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih C-J, Kao CM, Buttner MJ, Cohen SJ (2005) Cross regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol 58:1276–1287
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Inaoka T, Ochi K (2011) Scandium stimulates the production of amylase and bacilysin in Bacillus subtilis. Appl Environ Microbiol 77:8181–8183
Inaoka T, Takahashi K, Yada H, Yoshida M, Ochi K (2004) RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J Biol Chem 279:3885–3892
Iqbal M, Mast Y, Amin R, Hodgson DA, Wohlleben W, Burroughs NJ (2012) Extracting regulator activity profiles by integration of de novo motifs and expression data: characterizing key regulators of nutrient depletion responses in Streptomyces coelicolor. Nucleic Acids Res 40:5227–5239
Johansen SK, Maus CE, Plikaytis BB, Douthwaite S (2006) Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol Cell 23:173–182
Jornvall H, Hedlund J, Bergman T, Oppermann U, Persson B (2010) Superfamilies SDR and MDR: from early ancestry to present forms. Emergence of three lines, a Zn-metalloenzyme, and distinct variabilities. Biochem Biophys Res Commun 396:125–130
Kato JY, Funa N, Watanabe H, Ohnishi Y, Horinouchi S (2007) Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc Natl Acad Sci USA 104:2378–2383
Kawachi R, Akashi T, Kamitani Y, Sy A, Wangchaisoonthorn U, Nihira T, Yamada Y (2000) Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol Microbiol 36:302–313
Kawai K, Wang G, Okamoto S, Ochi K (2007) The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol Lett 274:311–315
Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC (2011) A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol 7:794–802
Kitani S, Yamada Y, Nihira T (2001) Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J Bacteriol 183:4357–4363
Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA J Am Med Assoc 298:1763–1771
Lahana R (1999) How many leads from HTS? Drug Discov Today 4:447–448
Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA 108:6258–6263
Lautru S, Challis GL (2004) Substrate recognition by nonribosomal peptide synthetase multi-enzymes. Microbiology 150:1629–1636
Lee J, Hwang Y, Kim S, Kim E, Choi C (2000) Effect of a global regulatory gene, afsR2, from Streptomyces lividans on avermectin production in Streptomyces avermitilis. J Biosci Bioeng 89:606–608
Lee PC, Umeyama T, Horinouchi S (2002) afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol Microbiol 43:1413–1430
Leipe DD, Landsman D (1997) Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res 25:3693–3697
Linares JF, Gustafsson I, Baquero F, Martinez JL (2006) Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci USA 103:19484–19489
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143
Liu M, Kirpekar F, Van Wezel GP, Douthwaite S (2000) The tylosin resistance gene tlrB of Streptomyces fradiae encodes a methyltransferase that targets G748 in 23S rRNA. Mol Microbiol 37:811–820
Maharjan S, Oh TJ, Lee HC, Sohng JK (2009) Identification and functional characterization of an afsR homolog regulatory gene from Streptomyces venezuelae ATCC 15439. J Microbiol Biotechnol 19:121–127
Malpartida F, Hopwood DA (1986) Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol Gen Genet 205:66–73
Manteca A, Fernandez M, Sanchez J (2005) A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology 151:3689–3697
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Martín JF (2004) Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol 186:5197–5201
Martín JF, Demain A (1980) Control of antibiotic biosynthesis. Microbiol Rev 44:230–251
Martin JF, Aparicio JF (2009) Enzymology of the polyenes pimaricin and candicidin biosynthesis. Methods Enzymol 459:215–242
Martin JF, Liras P (2010) Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 13:263–273
Martin JF, Sola-Landa A, Santos-Beneit F, Fernandez-Martinez LT, Prieto C, Rodriguez-Garcia A (2011) Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb Biotechnol 4:165–174
Martinez A, Kolvek SJ, Hopke J, Yip CL, Osburne MS (2005) Environmental DNA fragment conferring early and increased sporulation and antibiotic production in Streptomyces species. Appl Environ Microbiol 71:1638–1641
Matsumoto A, Ishizuka H, Beppu T, Horinouchi S (1995) Involvement of a small ORF downstream of the afsR gene in the regulation of secondary metabolism in Streptomyces coelicolor A3(2). Actinomycetologica 9:37–43
McDowall KJ, Thamchaipenet A, Hunter IS (1999) Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J Bacteriol 181:3025–3032
Medema M, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346
Medema MH, Breitling R, Bovenberg R, Takano E (2011) Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol 9:131–137
Mendes MV, Tunca S, Anton N, Recio E, Sola-Landa A, Aparicio JF, Martin JF (2007) The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab Eng 9:217–227
Moore JM, Bradshaw E, Seipke RF, Hutchings MI, McArthur M (2012) Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Methods Enzymol 517:367–385
Myers PL (1997) Will combinatorial chemistry deliver real medicines? Curr Opin Biotechnol 8:701–707
Nazari B, Kobayashi M, Saito A, Hassaninasab A, Miyashita K, Fujii T (2012) Chitin-induced gene expression involved in secondary metabolic pathways in Streptomyces coelicolor A3(2) grown in soil. Appl Environ Microbiol 79:707–713
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384
Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70:461–477
Nodwell JR, Losick R (1998) Purification of an extracellular signaling molecule involved in production of the aerial mycelium by Streptomyces coelicolor. J Bacteriol 180:1334–1337
Nothaft H, Dresel D, Willimek A, Mahr K, Niederweis M, Titgemeyer F (2003) The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J Bacteriol 185:7019–7023
Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F (2010) The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol 75:1133–1144
Novakova R, Rehakova A, Kutas P, Feckova L, Kormanec J (2011) The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 157:1629–1639
O’Rourke S, Wietzorrek A, Fowler K, Corre C, Challis GL, Chater KF (2009) Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol Microbiol 71:763–778
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97:87–98
Ochi K, Okamoto S (2012) A magic bullet for antibiotic discovery. Chem Biol 19:932–934
Ochi K, Tanaka Y, Tojo S (2013) Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol (submitted)
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Ohnishi Y, Kameyama S, Onaka H, Horinouchi S (1999) The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol 34:102–111
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA 98:12215–12220
Onaka H, Mori Y, Igarashi Y (2011) Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol 77:400–406
Paradkar A (2013) Clavulanic acid production by Streptomyces clavuligerus: biogenesis, regulation and strain improvement. J Antibiot. doi:10.1038/ja.2013.26
Parajuli N, Viet HT, Ishida K, Tong HT, Lee HC, Liou K, Sohng JK (2005) Identification and characterization of the afsR homologue regulatory gene from Streptomyces peucetius ATCC 27952. Res Microbiol 156:707–712
Pawlik K, Kotowska M, Chater KF, Kuczek K, Takano E (2007) A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch Microbiol 187:87–99
Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40
Piepersberg W, Distler J (1997) Aminoglycosides and sugar components in other secondary metabolites. In: Rehm HJ, Reed G (eds) Products of secondary metabolism, vol. 7, 2nd edn. VCH-Verlagsgesellschaft, Weinheim, pp 397–488
Pimm SL, Russell GJ, Gittleman JL, Brooks TM (1995) The future of biodiversity. Science 269:347–350
Qiu X, Yan X, Liu M, Han R (2012) Genetic and proteomic characterization of rpoB mutations and their effect on nematicidal activity in Photorhabdus luminescens LN2. PLoS One 7:e43114
Ratcliff WC, Denison RF (2011) Microbiology. Alternative actions for antibiotics. Science 332:547–548
Reading C, Cole M (1977) Clavulanic acid: a beta-lactamase-inhibiting beta-lactam from Streptomyces clavuligerus. Antimicrob Agents Chemother 11:852–857
Recio E, Colinas A, Rumbero A, Aparicio JF, Martín JF (2004) PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J Biol Chem 279:41586–41593
Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Muller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP (2006) The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol 61:1237–1251
Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP (2008) Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9:670–675
Rodriguez-Garcia A, Sola-Landa A, Apel K, Santos-Beneit F, Martin JF (2009) Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res 37:3230–3242
Romero D, Traxler MF, Lopez D, Kolter R (2011) Antibiotics as signal molecules. Chem Rev 111:5492–5505
Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, Sanchez M, Rocha D, Sanchez B, Avalos M, Guzman-Trampe S, Rodriguez-Sanoja R, Langley E, Ruiz B (2010) Carbon source regulation of antibiotic production. J Antibiot (Tokyo) 63:442–459
Santos-Beneit F, Barriuso-Iglesias M, Fernandez-Martinez LT, Martinez-Castro M, Sola-Landa A, Rodriguez-Garcia A, Martin JF (2011) The RNA polymerase omega factor RpoZ is regulated by PhoP and has an important role in antibiotic biosynthesis and morphological differentiation in Streptomyces coelicolor. Appl Environ Microbiol 77:7586–7594
Santos-Beneit F, Rodriguez-Garcia A, Sola-Landa A, Martin JF (2009) Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol 72:53–68
Sekurova O, Sletta H, Ellingsen TE, Valla S, Zotchev S (1999) Molecular cloning and analysis of a pleiotropic regulatory gene locus from the nystatin producer Streptomyces noursei ATCC11455. FEMS Microbiol Lett 177:297–304
Shapiro S (1989) Nitrogen assimilation in actinomycetes and the influence of nitrogen nutrition on actinomycete secondary metabolism. In: Shapiro S (ed) Regulation of secondary metabolism in actinomycetes. CRC Press, Boca Raton, pp 135–211
Sidda JD, Corre C (2012) Gamma-butyrolactone and furan signaling systems in Streptomyces. Methods Enzymol 517:71–87
Skinner R, Cundliffe E, Schmidt FJ (1983) Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem 258:12702–12706
Sola-Landa A, Moura RS, Martin JF (2003) The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc Natl Acad Sci USA 100:6133–6138
Sola-Landa A, Moura RS, Martín JF (2002) The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc Natl Acad Sci USA 100:6133–6138
Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64:435–459
Strauss J, Reyes-Dominguez Y (2011) Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol 48:62–69
Swiatek MA, Gubbens J, Bucca G, Song E, Yang YH, Laing E, Kim BG, Smith CP, van Wezel GP (2013) The ROK family regulator Rok7B7 pleiotropically affects xylose utilization, carbon catabolite repression, and antibiotic production in Streptomyces coelicolor. J Bacteriol 195:1236–1248
Swiatek MA, Tenconi E, Rigali S, van Wezel GP (2012) Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in the control of development and antibiotic production. J Bacteriol 194:1136–1144
Swiatek MA, Urem M, Tenconi E, Rigali S, van Wezel GP (2012) Engineering of N-acetylglucosamine metabolism for improved antibiotic production in Streptomyces coelicolor A3(2) and an unsuspected role of NagA in glucosamine metabolism. Bioengineered 3:280–285
Tamehiro N, Hosaka T, Xu J, Hu H, Otake N, Ochi K (2003) Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl Environ Microbiol 69:6412–6417
Tanaka Y, Hosaka T, Ochi K (2010) Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2). J Antibiot 63:477–481
Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K (2009) Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol 75:4919–4922
Titgemeyer F (2007) Carbon and nitrogen regulation in Gram-positive bacteria: a tribute to Milton H. Saier, Jr. J Mol Microbiol Biotechnol 12:5–8
Titgemeyer F, Reizer J, Reizer A, Saier MH Jr (1994) Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349–2354
Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS (2007) Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA 104:10376–10381
Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, McDowall KJ (2005) Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol Microbiol 58:131–150
van Wezel GP, Konig M, Mahr K, Nothaft H, Thomae AW, Bibb M, Titgemeyer F (2007) A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in Streptomyces coelicolor A3(2). J Mol Microbiol Biotechnol 12:67–74
van Wezel GP, Krabben P, Traag BA, Keijser BJ, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B (2006) Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl Environ Microbiol 72:5283–5288
van Wezel GP, McDowall KJ (2011) The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1333
van Wezel GP, McKenzie NL, Nodwell JR (2009) Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol 458:117–141
van Wezel GP, Titgemeyer F, Rigali S (2006) Methods and means for metabolic engineering and improved product formation by micro-organisms Patent application WO/2007/094667
van Wezel GP, White J, Hoogvliet G, Bibb MJ (2000) Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Mol Microbiol Biotechnol 2:551–556
Vigliotta G, Tredici SM, Damiano F, Montinaro MR, Pulimeno R, di Summa R, Massardo DR, Gnoni GV, Alifano P (2005) Natural merodiploidy involving duplicated rpoB alleles affects secondary metabolism in a producer actinomycete. Mol Microbiol 55:396–412
Vining LC (1992) Secondary metabolism, inventive evolution and biochemical diversity: a review. Gene 115:135–140
Vogtli M, Chang PC, Cohen SN (1994) afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol Microbiol 14:643–653
Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC (2012) Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA 109:E1743–E1752
WHO-Media-centre (2012) Antimicrobial resistance WHO.http://www.who.int/mediacentre/factsheets/fs194/en/
Wietzorrek AM, Bibb (1997) A novel family of proteins that regulates antibiotic production in Streptomyces appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1181–1184
Willey JM, Gaskell AA (2011) Morphogenetic signaling molecules of the streptomycetes. Chem Rev 111:174–187
Woodruff HB, Ruger M (1948) Studies on the physiology of a streptomycin-producing strain of Streptomyces griseus on proline medium. J Bacteriol 56:315–321
Xu Q, van Wezel GP, Chiu HJ, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Lesley SA, Godzik A, Elsliger MA, Deacon AM, Wilson IA (2012) Structure of an MmyB-like regulator from C. aurantiacus, member of a new transcription factor family linked to antibiotic metabolism in actinomycetes. PLoS One 7:e41359
Yamada Y, Nihira T (1999) Microbial hormones and microbial chemical ecology. In: Mori K (ed) Comprehensive natural products chemistry, vol. 8. Elsevier Scientific Publishers, Dordrecht, pp 377–413
Yamamoto S, He Y, Arakawa K, Kinashi H (2008) γ-Butyrolactone-dependent expression of the streptomyces antibiotic regulatory protein gene srrY plays a central role in the regulatory cascade leading to Lankacidin and lankamycin production in Streptomyces rochei. J Bacteriol 190:1308–1316
Yang YH, Song E, Willemse J, Park SH, Kim WS, Kim EJ, Lee BR, Kim JN, van Wezel GP, Kim BG (2012) A novel function of Streptomyces integration host factor (sIHF) in the control of antibiotic production and sporulation in Streptomyces coelicolor. Antonie Van Leeuwenhoek 101:479–492
Yang YL, Xu Y, Straight P, Dorrestein PC (2009) Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 5:885–887
Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S, Tan H, Lu Y, Jiang W (2012) Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol Microbiol 85:535–556
Zerikly M, Challis GL (2009) Strategies for the discovery of new natural products by genome mining. Chembiochem: Eur J Chem Biol 10:625–633
Acknowledgments
We are grateful to Young Choi for drawing chemical structures, to Geneviève Girard for comments on the manuscript, and to Kenneth McDowall, Marnix Medema and Michael Fischbach for sharing unpublished data. The work was supported by a CSC PhD fellowship from the Chinese government to HZ and by grant 10467 from the Netherlands Technology Foundation STW to GPvW.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special issue: Genome Mining for Natural Products Discovery.
Rights and permissions
About this article
Cite this article
Zhu, H., Sandiford, S.K. & van Wezel, G.P. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41, 371–386 (2014). https://doi.org/10.1007/s10295-013-1309-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1309-z