Abstract
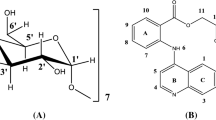
A lot of interest has been seen in computational methods that provide higher degree of accuracy and reliability, in structural determination and intermolecular interaction of different molecular systems. In the present work, atom accurate structure of β-cyclodextrin (β-CD) and midazolam (MDZ) was determined by a combination of NMR (2D-ROESY) and computational methods (MD) using quantitative approach, as developed and proved to be reliable by us previously. Further, we determined the structure of MDZ/β-CD inclusion complex by using molecular docking studies. Then, the structure obtained by molecular docking studies was compared with the previously developed method by quantitative approach. Parallel results were seen for the structure obtained by our method as well molecular docking approach. This validates the atom accuracy of the structures obtained by both MD and molecular docking.
Moreover, both the methods have advantages as well as disadvantages. The advantages of both the methods are that they are less time consuming and cheap. However, main disadvantage of MD is that final structure depends on initial frame and structure obtained from molecular docking depends on dimension of grid box and Cartesian coordinates. Therefore, both methods need to be performed several times to obtain atom accurate and reliable structures.









Similar content being viewed by others
Data availability
All the required data has been submitted to journal either in manuscript or as supplementary material.
References
Schneider HJ (1991) Mechanisms of molecular recognition: investigations of organic host–guest complexes. Angew Chem Int Ed Engl 30:1417–1436. https://doi.org/10.1002/anie.199114171
Safia H, Ismahan L, Abdelkrim G, Mouna C, Leila N, Fatiha M (2019) Density functional theories study of the interactions between host β-Cyclodextrin and guest 8-Anilinonaphthalene-1-sulfonate: molecular structure, HOMO, LUMO, NBO, QTAIM and NMR analyses. J Mol Liq 280:218–229. https://doi.org/10.1016/j.molliq.2019.01.019
Zaboub A, Madi F, Merdes R, Mohamedi M, Nouar L (2016) A combined DFT and experimental study of proline/β-cyclodextrin inclusion complex. J Mol Liq 216:716–723. https://doi.org/10.1016/j.molliq.2016.01.082
Bakirci HS, Zhang X, Nau WM (2005) Induced circular dichroism and structural assignment of the cyclodextrin inclusion complexes of bicyclic azoalkanes. J Org Chem 70:39–46. https://doi.org/10.1021/jo048420k
Raffaini G, Ganazzoli F, Malpezzi L, Fuganti C, Fronza G, Panzeri W, Mele A (2009) Validating a strategy for molecular dynamics simulations of cyclodextrin inclusion complexes through single-crystal X-ray and NMR experimental data: a case study. J Phys Chem B 113:9110–9122. https://doi.org/10.1021/jp901581e
Ali SM, Muzaffar S, Imtiaz S (2019) Comparative study of complexation between cyclodextrins and xylazine using 1H NMR and molecular modeling methods. J Mol Struct 1197:56–64. https://doi.org/10.1016/j.molstruc.2019.06.080
Lehmman J, Kleinpeter E (1991) 1H NMR spectroscopy as a probe of intermolecular interactions in β-cyclodextrin inclusion complexes. J Incl Phenom Mol Recognit Chem 10:233–239. https://doi.org/10.1007/BF01066207
Ali SM, Muzaffar S (2015) Quantitative ROESY analysis for unravelling structure of glafenine and β-cyclodextrin complex. J Incl Phenom Macrocycl Chem 94:95–102. https://doi.org/10.1007/s10847-019-00911-z
Muzaffar S, Imtiaz S, Ali SM (2020) Demonstrating accuracy of the proposed protocol for structure elucidation of cyclodextrin inclusion complexes by validation using DFT studies. J Mol Struct 1277:128419–128430. https://doi.org/10.1016/j.molstruc.2020.128419
Yahia HA, Yahia OA, Khatmi D, Belghiche R, Bouzitouna A (2017) Quantum chemical investigations on hydrogen bonding interactions established in the inclusion complex β-cyclodextrin/benzocaine through the DFT, AIM and NBO approaches. J Incl Phenom Macrocycl Chem 89:353–365. https://doi.org/10.1007/s10847-017-0753-1
Rafati AA, Hashemianzadeh SM, Nojini ZB, Safarpour MA (2007) Theoretical study of the inclusion complexes of α and β-cyclodextrins with decyltrimethylammonium bromide (DTAB) and tetradecyltrimethylammonium bromide (TTAB). J Mol Liq 135:153–157. https://doi.org/10.1016/j.molliq.2006.11.006
Yousuf I, Bashir M, Arjmand F, Tabassum S (2019) Multispectroscopic insight, morphological analysis and molecular docking studies of CuII based chemotherapeutic drug entity with human serum albumin (HSA) and bovine serum albumin (BSA). J Biomol Struct Dyn 37:3290–3304. https://doi.org/10.1080/07391102.2018.1512899
Ali SM, Fatma K, Dhokale S (2013) Structure elucidation of β-cyclodextrin-xylazine complex by a combination of quantitative 1H-1H ROESY and molecular dynamics studies. Beilstein J Org Chem 9:1917–1924. https://doi.org/10.3762/bjoc.9.226
Allinger NL (1977) Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms 1, 2. J Am Chem Soc. 99:8127–8134. https://doi.org/10.1021/ja00467a001
Ali SM, Shamim S (2015) Quantitative ROESY analysis of computational models: structural studies of citalopram and β-cyclodextrin complexes by 1H-NMR and computational methods. Magn Reson Chem 53:526–535. https://doi.org/10.1007/s10847-019-00911-z
Kohler JEH, Grczelschak-Mick N (2013) The β-cyclodextrin/benzene complex and its hydrogen bonds – a theoretical study using molecular dynamics, quantum mechanics and COSMO-RS, Beilstein J Org Chem 9:118-134. https://doi.org/10.3762/bjoc.9.15
Jingye L, Deyue Y, Qun C (2002) Preparation and characterization of the crystalline inclusion complexes between cyclodextrins and poly(1,3-dioxolane), Sci China Ser B 45:73-83. https://doi.org/10.1360/02yb9011
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. https://doi.org/10.1021/ci3001277
Eddine ZK, Fatiha M, Amal Z, Leila N, Rachid M (2015) Investigation of the inclusion complex of tolfenamic acid with β-cyclodextrin: Geometry and NBO analysis. Comptes Rendus Chimie. 18:193–199. https://doi.org/10.1016/j.crci.2014.04.012
Butts CP, Jones CR, Towers EC, Flynn JL, Appleby L, Barron NJ (2011) Interproton distance determinations by NOE-surprising accuracy and precision in a rigid organic molecule. Org Biomol Chem. 9:177–184. https://doi.org/10.3762/bjoc.7.20.
Grube A, Kock M (2007) Structural Assignment of Tetrabromostyloguanidine: Does the Relative Configuration of the Palauamines need revision? Angew Chem Int Ed. 46:2320–2324. https://doi.org/10.1002/anie.200604076.
Chini MG, Jones CR, Zampella A, Auria MVD, Renga B, Fiorucci S, Butts CP, Bifulco G (2012) Quantitative NMR-Derived Interproton Distances Combined with Quantum Mechanical Calculations of 13C Chemical Shifts in the Stereochemical Determination of Conicasterol F, a Nuclear Receptor Ligand from Theonella swinhoei. J Org Chem. 77:1489–1496. https://doi.org/10.1021/jo2023763.
Acknowledgments
Shah Imtiaz and Sughra Muzaffar are thankful to UGC for providing financial support. We are also thankful to Department of chemistry for providing basic facilities for research.
Contributor information
Shah Imtiaz, Email. Shahimtiaz3195@gmail.com
Syqa Banoo, Email shahimtiazphd@gmail.com
Sughra Muzaffar, Email. muzaffarsughra@gmail.com
Syed Mashhood Ali, Email. smashhoodali@gmail.com
Author information
Authors and Affiliations
Contributions
Shah Imtiaz, Syed Mashhood Ali, and Syqa Banoo designed experiment, interpreted data, wrote manuscript. Sughra Muzaffar wrote manuscript, revised manuscript with minor changes. Syed Mashhood Ali approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent to publish
The authors declare that they show their consent to publish.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 39 kb)
Rights and permissions
About this article
Cite this article
Imtiaz, S., Banoo, S., Muzaffar, S. et al. Structural determination of midazolam/beta-cyclodextrin inclusion complex by an already proposed protocol and molecular docking studies by quantitative analysis. Struct Chem 32, 1505–1516 (2021). https://doi.org/10.1007/s11224-021-01727-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-021-01727-9