Abstract
Aims
The phyllosphere and soil are crucial and distinct microbial spheres in forests, connected through trees that interact with both. As part of the tree's holobiont, these communities are vital to the fitness and evolution of the host. Differences between the spheres may be particularly evident at the two extreme ends of tall and long-lived trees of natural temperate forest; the top-canopy and the soil. Here, we evaluated the connectivity between the top-canopy and soil microbial communities of European beech and Norway spruce trees to determine the significance of tree-sphere and host-species identity, and to assess the contribution of taxa inhabiting both spheres.
Methods
Bacterial and fungal community composition was determined through metabarcoding analysis of linked top-canopy leaf and bulk soil samples collected from tall (old) trees in the natural forest of Bavarian Forest National Park.
Results
This study shows sphere-specific communities in European temperate forests, characterized by low connectivity. Results highlight that spheres exert stronger influence than host identity. Only a few taxa inhabited both spheres, yet they accounted for the bulk of the (relative) abundance in each sphere.
Conclusion
Analysing the divergence and shared characteristics of these interlinked communities redefines the tree holobiont concept and enhances our understanding of the evolution of plant-associated microbial communities in a sphere-specific manner. This study emphasizes the importance of examining multiple microbiome components for a thorough understanding of temperate forest ecology, while also highlighting the existence of a small group of overlapping taxa that may play a bigger role than previously anticipated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Temperate forests are structurally complex ecosystems containing a great variety of habitats. Biotic and abiotic variables exhibit a high level of spatial heterogeneity within forest ecosystems, both horizontally (e.g., between different forest stands) and vertically, from the soil to the top canopy. The microbial communities living in these habitats play important ecological roles at multiple spatial scales (Baldrian 2017). They impact individual plant health as either plant-promoters or pathogenic taxa (Terhonen et al. 2019), and also provide ecosystem services due to their role in, among others, nutrient cycling, and carbon sequestration (Graham et al. 2016). Forest microbial communities generally display high diversity, encompassing both r-strategists (higher growth-rates in resource-rich environments) and K-strategists (slow growth but adept energy source utilization) (MacArthur and Wilson 1967). These communities can be can be shaped by both deterministic (where species occurrence and abundance stem primarily from abiotic and biotic factors) and stochastic processes (influenced by random and probabilistic events) (Dini-Andreote et al. 2015). Soils host some of the densest microbial communities on earth, with high taxonomical and functional diversity (Pulleman et al. 2012). Forest soils provide a vast habitat for microbes, being characterised by high nutrient levels and extensive spatial heterogeneity of micro-habitats (Kadowaki et al. 2014; Martins et al. 2013; Zhou et al. 2022). The canopy also covers a massive surface area which is characterized by an oligotrophic environment in combination with rapidly fluctuating environmental stressors such as ultraviolet (UV) radiation and desiccation (Lindow and Brandl 2003). The soil and canopy are thus important “spheres” in forest ecosystems, depicting the extremities of tall and long-lived trees. In the context of plants, we define sphere as a term to describe the zone of influence or the area of interaction between the plant, environmental components, and microbial communities. Examples of plant spheres are the phyllosphere (the microenvironment on aerial plant surfaces), rhizosphere (the soil zone surrounding/adhering to a root), endosphere (habitats within the plant's tissues) and the spermosphere (zone around the seeds of a plant) (Bais et al. 2006; Lemanceau et al. 2017; Vacher et al. 2016). The host plants connect these different spheres, forming a complex ecological unit with the microbial communities living within these spheres. The plant, as host organism, and its associated symbiotic microbial community are often considered as a holobiont in which the microbes play a central role in host biology, ecology, and evolution, and vice versa (Simon et al. 2019).
The concept of the holobiont emphasizes the idea that host organisms and their associated microorganisms are interdependent and co-evolve over time (Zilber-Rosenberg and Rosenberg 2008). Plants actively select microbial communities via plant exudation and anatomical properties (Whipps et al. 2008; Zhou et al. 2022). The plant is also a prominent factor in the (cross-)colonization of the different spheres, for example via xylem and phloem transport, leaf fall or seed germination (Barret et al. 2015; Chi et al. 2005; Guerreiro et al. 2017). Other colonization pathways include transport via air, rainfall/stemflow, rain splash or organisms (insect, bird, animal) (Bittar et al. 2018; Coluccio et al. 2008; Levetin and Dorsey 2006; Zarraonaindia et al. 2015; Zhou et al. 2020). In the soil, trees can influence microbial communities up to several meters from the tree stem, as shown by studies on soil microbial communities under European beech and Norway spruce trees (Nacke et al. 2016). Within the tree-associated soil habitat, rhizosphere communities can be considered as a subset of bulk soil communities (bare soil next to the plant/roots), with the latter showing a lower density but higher diversity of microbes (Bulgarelli et al. 2013; Zhou et al. 2022). The rhizosphere has, compared to the bulk soil, a limited spatial influence beyond immediate adjacency to the direct fine root material since it only covers nanometers or milimeters from the roots.
Analysing overlapping taxa and connectivity between the different spheres of a plant can provide essential insight in plant health, holobiont evolution, and ecosystem response to environmental change (Coince et al. 2014; Zilber-Rosenberg and Rosenberg 2008). Microbial communities do not solely affect processes within a sphere but can also exert effects throughout various plant compartments, either by influencing plant health (i.e., via biomass, metabolites or pathogen resistance) (Berlanga-Clavero et al. 2020) or by microbe-microbe interactions via the secretion of secondary metabolites (Liu et al. 2017). While different spheres can be controlled by dissimilar environmental variables resulting in divergent communities (Coince et al. 2014), abundant generalist microbes occurring in multiple spheres may be part of a core microbiome; being on the one hand actively selected by the host plant (Bai et al. 2022) and on the other hand shaping microbe-microbe and microbe-plant interactions throughout the host (Hassani et al. 2018). Via these interactions, these shared taxa may play a central role in host ecology, fitness and evolution (Simon et al. 2019). Studies on crops and annuals show varying levels of similarities and connectivity between the above-ground and below-ground plant associated communities (Bai et al. 2015; Tkacz et al. 2020; Zarraonaindia et al. 2015), but information about the connectivity of different spheres in natural temperate forests is scarce (some relevant examples: Coince et al. 2014; Haas et al. 2018; Rodríguez-Rodríguez et al. 2023). The little research on the connectivity of these spheres in European forests is often either experimental (e.g. Haas et al. 2018; Potthast et al. 2022) or focuses on a small number of plants or plots (e.g. Beule et al. 2017; Coince et al. 2014). Connectivity and taxonomic similarities between these different microbial communities may be especially complex in trees due to the tree’s longevity and large size, resulting in sphere-specific variation in environmental stresses and host-interactions (Flessa et al. 2012; Herrmann et al. 2021), calling for a deeper comprehension of the overlap and divergence within the tree holobiont.
In the current paper, we analyse and compare the bacterial and fungal communities in the canopy and soil of the temperate forests of Bavarian Forest National Park (Germany), in order to assess their connectivity and divergence. We will focus on the communities located in top canopy and tree-associated bulk soil (i.e., under the canopy and in the zone of influence of the individual trees; Nacke et al. 2016) since these spheres are located at the most extreme, and environmentally dissimilar ends of the tree holobiont. Our hypotheses are: (i) Variation in European beech and Norway spruce leaf and soil bacterial and fungal communities can be primarily attributed to tree sphere rather than host-species identity. (ii) Taxa possessing the capability to inhabit both spheres demonstrate a competitive advantage, leading to heightened (relative) abundances. Insights gained on the complex interactions of these distinct microbiomes have high relevance for developing strategies to manage and conserve forest ecosystems. Our study highlights the necessity to incorporate microbial diversity indices of multiple spheres in ecosystem assessments since this will provide a more all-inclusive picture of variation and vulnerability of microbial communities and ecosystem functions in European temperate forests.
Methods
Site description and sampling design
Top-canopy and bulk soil samples were collected in twenty-nine square plots of 30 × 30 m in Bavarian Forest National Park (Fig. 1). The park is located in south-eastern Germany and is part of the Bohemian Forest, one of the most extensive contiguous natural forest ecosystems in Central Europe (Křenová and Kiener 2013). The mountainous forests in the study area (altitudes between 300–1400 m a.s.l.) are dominated by European beech (Fagus sylvatica) and Norway spruce (Picea abies) (Bässler et al. 2015). The park was established in 1970 and has a 40-year no-intervention management strategy (since 1983), resulting in a gradual decline in spruce (mainly due to bark beetle infestations) and an increase in abundance of beech (as well as, to some extent, other tree species such as Abies alba) (van der Knaap et al. 2020).
Plots were stratified over stands (> 75% canopy dominance) of European beech (N = 15) and Norway spruce (N = 14), and were situated at altitudes ranging from 665 to 1160 m. Sampling was conducted in the same season, during July–August 2020, while broad leaves were fully mature and before senescence (Laforest-Lapointe et al. 2016). At each 300 m2 plot, leaves were collected from three representative trees. Per tree, a leaf sample consisted of 10 individual broadleaves from different branches or 20–30 needles from 10 separate cohorts (internodes of the same age). Leaf samples were collected from the sun-exposed top canopy using a large slingshot (Tree runner BigShot) and a rope (Youngentob et al. 2016). A modified crossbow was used for tall trees (Ali et al. 2016). Wearing disposable gloves, falling leaves were caught before touching the ground to avoid contamination and stored in zip-lock bags. Leaves were pooled per tree to reduce between-leaves variation (Cordier et al. 2012). Per tree, a composite bulk soil sample was collected from the topsoil (0–10 cm depth, after removal of litter) in a 9 m2 subplot located under the canopy of the sampled tree (max 2 m distance from the tree trunk). Per subplot, 9 cores were collected in a 3 × 3 grid using a 5 cm Ø × 5 cm height corer. Composite samples were collected by pooling the nine cores in a sterile bag, removal of roots and stones, manual homogenization, and subsequently transferring a subsample into sterile 50 ml tube. Samples were transported on ice to the laboratory where they were stored at -20 °C until further processing. Between samples, the soil corer was sterilized with 10% bleach followed by deionised water to avoid cross-contamination. For each fifth plot, an aliquot of the deionised water rinse of the corer was collected as field control.
DNA extraction and amplification
A sterile paper hole puncher (0.6 cm Ø) was used to punch leaf disks from broadleaf samples. For each tree, 0.1 g of broadleaf leaf-disks or needles were combined (representing 10 broadleaves or 10 needle cohorts) and homogenized using a Benchmark Beadbug™ Mini Homogenizer (D1030). Leaf-disks were cut-out and needles were selected from different parts of the leaves/cohorts to warrant a representative sample. Leaf total DNA (endophytes and epiphytes) was extracted using Qiagen Plant Pro extraction kit and the Qiagen Qiacube Connect extraction robot, following the manufacture’s instructions. No sterilization was conducted prior to DNA extractions; total DNA from combined endophyte and epiphyte taxa was extracted (Zarraonaindia et al. 2015). Soil DNA was extracted following the phosphate extraction protocol of Taberlet et al. (2012), using minor modifications. Briefly, 15 g of well-homogenised soil sample was mixed with 15 ml saturated phosphate buffer (Na2HPO4; 0.12 m; pH ≈8) in order to extract extra-cellular DNA. After mixing and centrifugation, 2 mL of supernatant was purified using the NucleoSpin® soil extraction kit following the manufacture’s instructions, but omitting the lysis step. Using the saturated phosphate buffer enabled processing larger soil volumes compared to the kit's lysis step, minimizing the effects of local heterogeneity. Negative extraction controls were included for each batch of 22 (leaf) or 25 (soil) samples. DNA concentrations were quantified on a Biotek Synergy HTX Multi Mode Reader, using the Quant-iT PicoGreen dsDNA Assay Kit, and standardised to 5 ng µl−1 (samples < 5 ng µl−1 were not standardised). Soil samples were further 100 × diluted to reduce polymerase chain reaction (PCR) inhibition. Prior to PCR, the field and extraction controls for each sample and control type were combined.
DNA extraction and amplification were conducted at separate laboratories to reduce contamination risks. Bacterial (16S rRNA gene) and fungal (ITS rRNA region) DNA was amplified using the 515F/806R (Apprill et al. 2015; Parada et al. 2016) and ITS86/ITS4-ngs (Tedersoo et al. 2014; Turenne et al. 1999) primer sets, respectively. Amplification protocols and polymerase chain reaction (PCR) recipes are shown in supplementary table 1. Peptide Nucleic Acid (PNA) clamps were used (PNA Bio Inc.) to block the amplification of host chloroplast and mitochondrial DNA in the leaf 16S PCR reactions (Lundberg et al. 2013). Amplicons were sent to Genome Quebec (Montreal, Canada) for library preparation and Next Generation paired-end sequencing. Primers contained a CS1 (forward primer) or CS2 (reverse primer) adaptor sequence at the 5’-end to allow for multiplexing using the Fluidigm Access Array System (Fluidigm, South San Francisco, CA). An indexing PCR was used to attach the indexes and i5/i7 Illumina adapter sequences to the amplicons. Sequencing was performed on one lane of the Illumina NovaSeq 6000 SP platform using the PE250 kit.
Bioinformatic and data analyses
Bioinformatic analyses were performed using the QIIME 2™ software suite (Bolyen et al. 2019) and all statistical analyses were performed in R version 4.2.3 (https://www.R-project.org/). Post-clustering curation was conducted using LULU (Frøslev et al. 2017), and the SILVA (Quast et al. 2012) and UNITE (Nilsson et al. 2018) databases were used for Taxonomical assignment. ASV tables were further (i) blank corrected (removal criteria: max reads in blanks ≥ max reads in samples), (ii) filtered to retain only bacterial and fungal reads, and (iii) corrected for tag-switching (following: Taberlet et al. 2018). All taxa present with less than 10 reads in total were removed to reduce low frequency noise (Alsos et al. 2016; Polling et al. 2022). Details of the bioinformatic pipeline can be found in supplementary table 2. Curated ASV tables were rarefied to 15,000 reads (supplementary Fig. 1) using the ‘rrarefy’ function of the Vegan v. 2.6–2 R package (Oksanen et al. 2022). The average of 100 rarefactions was used to reduce stochastic effects (Cordier et al. 2019). The leaf and soil sample pairs of two subplots/trees were removed from the ITS dataset due to a read count lower than 15,000, and one plot only contained information of 2 trees/subplots. The package ‘Vegan’ was also used to visualise and test for differences in community composition between spheres (leaf vs. soil samples) and tree species, using Principle Coordinate Analyses (PCoA) and PERMANOVA (999 permutations) on Bray–Curtis dissimilarities constructed from Hellinger-transformed read counts. Pearson correlations between communities were tested using Mantel tests. Permutations were restricted per plot to account for within plot pseudo-replication in PERMANOVA and Mantel tests. Differences in alpha diversity were visualised using ASV accumulation curves and tested using linear mixed models (‘nlme’, v. 3.1–162) (Pinheiro et al. 2022) using plot ID as random effect. Venn diagrams were constructed (‘ggvenn’ v. 0.1.9) (Yan 2021) to visualise differences in ASV overlap between spheres and tree species and differences in the fraction of ASVs overlapping per sample were determined using linear mixed models (random effect: plot ID) on logit-transformed proportions. Connectivity between the spheres was assessed using SPIEC-EASI (Sparse InversE Covariance estimation for Ecological Association and Statistical Inference) co-occurrence networks (Kurtz et al. 2015) incorporated in the package ‘NetCoMi’ (Peschel et al. 2020). Soil and leaf data were combined per tree to allow for the construction of across-sphere networks, and taxa occurring in both spheres were allocated multiple nodes for each respective sphere. Data were filtered to remove any ASVs with < 10% prevalence, and read counts were centered log-ratio (clr) transformed as part of the SPIEC-EASI algorithm. Networks were constructed and compared using the ‘NetCoMi’ package, and the adjacency matrix was used to classify edges as within-spheres or between-spheres associations. Hub taxa (“key-species”) were assigned based on highest eigenvector centralities (> 95% quantile), representing nodes that have a central position in the network (Peschel et al. 2020). Discriminant analyses (‘Maaslin2’ v. 1.12) (Mallick et al. 2021) on Hellinger-transformed read counts were ran, with Plot ID as random effect, to determine sphere specificity of bacterial and fungal families. Families were classified as unique (occurrence in only one sphere), dominant (discriminant [p < 0.05] to a specific sphere), or non-specific (not discriminant [p > 0.05] to a specific sphere). Differences in relative read abundances of discriminant taxa (ASVs with > 10% prevalence) between spheres and tree species were further visualised over multiple taxonomic levels using heat trees analyses (‘metacoder’ v. 0.3.5) (Foster et al. 2017).
Results
Data summary
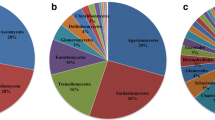
A total of 31,620,621 bacterial and 34,032,206 fungal reads remained after bioinformatic processing (supplementary table 2), with a mean (± SD) read count of 92,089 (± 42,250) 16S rRNA and 147,280 (± 86,560) ITS rRNA in leaf and 275,592 (± 41,831) 16S rRNA and 248,443 (± 36,380) ITS rRNA in soil samples. Rarefaction to 15,000 reads per sample resulted in a total of 38,538 bacterial ASVs (mean ± SD leaf: 389 ± 194; mean ± SD soil: 2235 ± 507) and 5,168 fungal ASVs (mean ± SD leaf: 246 ± 96; mean ± SD soil: 101 ± 33), al loss of 856 bacterial (2.2%) and 62 Fungal (1.2%) ASVs compared to the non-rarefied dataset. Phyllosphere bacterial communities were dominated by Proteobacteria (mean relative abundance in European beech samples: 55%; Norway spruce samples: 58%), Bacteroidota (beech: 34%; spruce: 9%) and Acidobacteriota (beech: 5%; spruce: 18%) while soil bacterial communities were dominated by Acidobacteriota (beech: 33%; spruce: 35%), Proteobacteria (beech: 22%, spruce: 19%) and Actinobacteriota (beech: 11%, spruce: 16%). Phyllosphere fungal communities mainly consisted of Ascomycota (beech: 97%, spruce: 93%) while the most abundant phyla in soil fungal communities were Basidiomycota (beech: 77%, spruce: 71%) and, to a lesser extend, Ascomycota (beech: 22%, spruce: 27%).
Diversity and community composition differences between compartments and host tree species
Bacterial and fungal communities differ both between plant spheres and host tree species (Fig. 2). PERMANOVA and Principle Coordinate (PCoA) analyses showed a significant (p < 0.01) influence of sphere and tree species, and a significant (p < 0.01) interaction between them for both 16S and ITS (Fig. 2; supplementary table 3). Samples clustered among spheres along the first PCoA axis, which explained 40% and 21% of the variance in community structure for 16S and ITS respectively. Clustering based on tree-species was along the second PCoA axis for the leaf samples (explaining 11% and 14% of the variance for 16S and ITS respectively) and the third axis for the soil samples (explaining 6% and 5% of the variance for 16S and ITS respectively). PERMANOVA analyses (supplementary table 3) confirmed the attribution of a larger proportion of variation to tree species in leaf samples (bacterial R2: 0.34; fungal R2: 0.39) than in soil samples (bacterial R2: 0.06; fungal R2: 0.09).
Total bacteria richness was several orders of magnitude higher in soil samples than in leaf samples and only the ASV accumulation curves of leaf samples showed no overlap in 95% confidence intervals between tree species (Fig. 3A). Total fungal richness was a factor 10 lower compared to total bacterial richness, which could mainly be attributed to a substantially lower diversity detected in the soil. Differences in fungal richness between spheres varied per tree species (Fig. 3B). Linear mixed model analyses of Shannon diversity on the subplot level (Fig. 3C-D) confirmed these patterns with significant differences between groups for both bacterial (F83 = 524.8, p < 0.0001, marginal R2 [R2m] = 0.80) and fungal (F81 = 54.9, p < 0.0001, R2m = 0.52) ASVs. Pairwise-comparisons showed significantly (p < 0.001) higher bacterial Shannon diversity in soil versus leaf samples and beech leaf versus spruce leaf samples, while soil bacterial diversity did not differ significantly (p = 0.12) between tree species (Fig. 3C). Spruce leaf samples contained a significant (p < 0.0001) higher fungal Shannon diversity than beech leaf samples and soil samples, independent of tree species. Soil fungal diversity did not differ significantly between tree species (Fig. 3D; p = 0.19).
Variation in bacterial (A-C) and fungal (B-D) accumulative ASV richness (± 95% confidence interval) and mean Shannon diversity of leaf and soil samples (rarefied to 15,000 reads) collected from two tree species (European beech and Norway spruce). Different letters indicate significantly different groups (linear mixed model with Benjamini–Hochberg pairwise comparisons, p < 0.05)
ASV connectivity between phyllosphere and soil
The majority of bacterial and fungal ASVs were unique to either the soil or to the leaf samples (Fig. 4 & supplementary table 4). A substantially lower proportion of families was unique to either of the spheres compared to ASV-level taxa specificity (supplementary table 4), indicating that most variation in unique taxa is at lower taxonomic ranks. Only a small proportion of ASVs occurred in both spheres (bacterial: 2.8%; fungal: 6.0%). See supplementary table 5 for the top 10 overlapping taxa per kingdom. A total of 448 bacterial ASVs were detected in both soil and leaf samples within the same subplot, with a significantly higher mean proportion of shared bacterial ASVs in spruce compared to beech subplots (LMM: F27 = 22.4, p < 0.001, R2m = 0.23). Although when comparing proportions separately per sphere, this difference was only significant for the soil communities (supplementary Fig. 2A-C). The number of fungal ASVs detected in both soil and leaf samples within the same subplot was 119, and no significant difference was detected in the mean proportion of shared fungal ASVs per subplot between tree species (supplementary Fig. 2D-F; LMM: F27 = 1.52, p = 0.23, R2m = 0.02). A significant positive correlation was found in the community composition of bacterial phyllosphere and soil communities in Norway spruce forests (Mantel test: R = 0.32 P < 0.01). No significant correlations, though, were detected in European beech forest communities (supplementary table 6). Also no correlations were shown for Alpha diversity between the leaf and soil samples, independent of tree species or microbial kingdom (supplementary table 6). Correlation analyses of the relative abundance of individual families between spheres did only detect three families with significant (BH-adjusted) Spearman correlations in Norway spruce forest stands (Xanthobacteraceae: R = 0.56, P < 0.05; Pleomassariaceae: R = 0.56, P < 0.01; and Sclerococcaceae: R = 0.64, P < 0.001), while none of the bacterial or fungal families showed significant correlations between the two spheres in European beech forest stands.
Co-occurrence network analyses showed no explicit clustering of taxa (nodes) per sphere in either the bacterial nor the fungal networks (Fig. 5). Global network statistics (supplementary table 7) showed differences between bacterial and fungal networks, especially in terms number of nodes (2068 vs. 142), modularity (0.2 vs. 0.6) and positive edge percentages (57–58% vs. 64–74%), indicating kingdom specific variation in ecological associations within the host tree. Fungal networks showed more pronounced differences in network structure between host tree species compared to bacterial networks. Particularly the number of components (48 vs. 16), clustering coefficient (0.13 vs. 0.07) and positive edge percentage (74% vs. 64%) were higher in the fungal beech network than the fungal spruce network. Taxa detected in soil samples were dominant in the bacterial networks, as shown by the high number of nodes and hubs (i.e. keystone species), while the fungal networks were dominated by taxa from leaf samples. The low number of edges (6–12%) between taxa from different spheres indicates low connectivity between the spheres (Table 1). Even for the few taxa that do occur in both spheres, the nodes were not linked, i.e. occurrence patterns did not correlate between spheres.
Co-occurrence networks of bacterial (A-B) and fungal (C-D) ASVs (> 10% prevalence) detected in forest bulk soil and phyllosphere samples. Data from soil and leaf samples was combined per tree prior to SPIEC-EASI network construction, and taxa occurring in both spheres were allocated multiple nodes for each respective sphere. Edge colour represents negative (red) and positive (green) associations. Hub nodes (based on eigenvector centralities) have a red outline. Node size represents differences in relative read abundance. Maximum 1000 nodes with the highest degree are shown
Taxa specificity and relative abundance in phyllosphere and soil
Even though the majority of ASVs detected were unique to either the soil or the leaf samples (Fig. 6A), the contribution of these unique taxa was low in terms of relative abundance (Fig. 6B). Discriminant taxa (significant higher relative abundance in leaf or soil samples based on Maaslin2 discriminant analyses) showed a substantially lower ASV richness compared to unique taxa but contributed most to the microbial communities in terms of relative read abundance, especially in leaf samples (Fig. 6). Variation in taxa specificity between tree species was most pronounced in the phyllosphere. Spruce stands show a higher contribution of unique leaf bacterial and fungal ASVs, both in terms of richness and relative abundance (Fig. 6). Soil profiles did not show pronounced differences in the ratio unique:dominant taxa of both microbial kingdoms, either in terms of richness or relative abundance (Fig. 6). The phyllosphere communities also showed clear patterns in variation in the relative abundance of leaf-discriminant taxa between the beech and spruce phyllosphere, while this was much less apparent in the soil (Fig. 7, supplementary Figs. 3 and 4).
Variation in ASV specificity of bacterial (A & B) and fungal (C & D) detected in leaf and soil samples in European beech and Norway spruce forests stands. Panels A and C show variation in ASV richness and panels B and D show variation in mean relative read abundances. Samples have been rarefied to 15,000 reads and specificity was determined by Maaslin2 discriminant analyses
Heat trees summarising differences in relative read abundance of bacterial (A-B) and fungal (C-D) taxa detected in soil and leaf samples over different taxonomic levels. Nodes highlighted in brown and green were significantly (p < 0.05) discriminant for the soil and leave compartments, respectively (based on Maaslin2 discriminant analyses). Colour intensity represent differences in Log2 ration median proportions of relative read abundances and node size represents total read count of the taxon. Abundant families (> 1% mean relative abundance) are highlighted. Full taxonomical key is provided in supplementary Figs. 3–4. Samples were rarefied to 15,000 reads prior to analysis
Sphere-specific patterns of dominant taxa
Abundant families mainly varied in terms of relative abundance between spheres (Fig. 8). Soil-discriminant bacteria were found in a wide range of phyla (Fig. 8A) while soil-discriminant fungi mainly belonged to one phylogenetic group, the order Agaricomycetes in the phylum Basidomycota (Fig. 8B). In case of leaf-discriminant taxa, both the bacterial and fungal taxa belonged to a wide range of phylogenetic groups (Fig. 8). Acidobacteriota were underrepresented in the phyllosphere with Acidobacteriaceae as notable exception (Fig. 8A). Abundant (> 5%) ASVs in the leaf samples were assigned to genera 1174–901-12 (Beijerinckiaceae) and Mucilaginibacter while the most abundant (> 1%) ASVs in the soil samples were all uncultured Acidobacteriales (supplementary table 8). Abundant (> 5%) fungal ASVs in the leaf samples were assigned to the genera Naevala and Erysiphe, and the family Phaeosphaeriaceae. In the soil samples, the most abundant (> 5%) fungal taxa were Russula cyanoxantha, Russula vesca, Imleria badia and an unassigned ASV in the order Atheliales (supplementary table 8).
Heatmaps of bacterial (A) and fungal (B) taxa identified in phyllosphere and soil samples in European beech and Norway spruce forest stands, showing variation in mean relative read abundance and prevalence (proportion of samples present) of abundant families (> 1% mean relative abundance). Family specificity was determined by Maaslin2 discriminant analyses and samples were rarefied to 15,000 reads prior to analysis
Discussion
The top canopy phyllosphere and bulk-soil bacterial and fungal communities of European beech and Norway spruce trees in Bavarian Forest National Park showed distinct microbial communities, with little overlap. In line with our first hypothesis, tree spheres had a multitude stronger (2–7 times) influence than host-species on both the fungal and bacterial communities, respectively, indicating that the microbiome ‘worlds’ in the soil under a tree and in the leaves of the upper crown are detached in terms of community composition, both across tree species as well as within a host species.
Host identity effects were sphere and kingdom specific, with main differences between beech and spruce fungal communities in the phyllosphere. While the majority of taxa were rare and specialised in either the soil or top-canopy, the few taxa possessing the capability to inhabit both spheres were relatively abundant, as postulated in our second hypothesis. Divergent microbial communities between plant spheres have been shown in a large variety of plant species (Fonseca-García et al. 2016; Yang et al. 2022), including a limited number of studies on temperate European natural forests (e.g. Beule et al. 2017; Coince et al. 2014; Haas et al. 2018; Lynikiene et al. 2020; Schneider et al. 2021). Our study adds to this knowledge by providing a comprehensive representation of the microbial communities across multiple spheres of the same individual tall forest tree, revealing patterns in two environmental spheres and among two microbial kingdoms sampled from 86 trees distributed over 29 plots in one of the largest contiguous natural forest ecosystems in Central Europe (Křenová and Kiener 2013). By sampling the top-canopy and bulk soil, our study focusses on the two most extreme environmental spheres of the tree’s holobiont which have distinct functions in the forest ecosystem (Bulgarelli et al. 2013).
Limited connectivity and sphere-specific community composition
The phyllosphere and soil are two unique ecological spheres that are linked by the same host plant (Vandenkoornhuyse et al. 2015). The host plant and associated microbial communities can be viewed as an holobiont in which the microbes play a central role in host biology, ecology, and evolution (Simon et al. 2019). The host tree has been traditionally postulated to be an important constant in this assemblage, not only by physically connecting the different microbial components, but also by actively selecting and shaping these communities by selective pressure via root exudates, secondary metabolites, and leaf chemical and topological properties (Bais et al. 2006; Cesarz et al. 2013; Karamanoli et al. 2005). Our results, however, indicate that the environmental sphere has a larger influence on tree-associated communities than host-selective pressures. Additionally, network analyses showed limited connectivity between the spheres, with only a fraction (6–12%) of associations detected across spheres. Concerning these few co-occurrences that span across spheres, it is improbable that they originated from direct interactions among microbiome species. Instead, they are more likely the outcome of indirect interactions facilitated through the host tree. Known examples of these indirect interactions within host plants are the transfer of genes, fluctuations in the host’s C-dynamics and the production of phytohormones (Chen et al. 2017; Liu et al. 2017; Potthast et al. 2022). The bacterial network exhibited a substantial contribution of taxa from soil samples, both in terms of number of nodes and hub taxa, while their role in the fungal network was comparatively minor. A similar pattern was observed in the heat trees, with soil-specific taxa dominating the bacterial trees and leaf-specific taxa being most apparent in the fungal trees. The leaf-taxa dominated fungal networks also showed a 3.8 times higher modularity compared to the soil-taxa dominated bacterial networks, indicating a higher subdivision of the network in different subcommunities which potentially inhabit different niches within the phyllosphere (Abdelfattah et al. 2016; Herrmann et al. 2021). Soil environments provide temporally stable, highly nutritious and micro-heterogenous habitats while the phyllosphere is characterised by oligotrophic habitats with harsher environmental conditions and large temporal variation (Kadowaki et al. 2014; Lindow and Brandl 2003; Zhou et al. 2022). As a result of these sphere-specific environmental influences, forest soil communities are driven by long-term and deterministic processes, such as long-term forest stand conditions and micro-scale variation in soil conditions (especially pH) (Hannam et al. 2007; Rodríguez-Rodríguez et al. 2023), while phyllosphere communities are largely influenced by host-identity (Laforest-Lapointe et al. 2016) and spatio-temporal variation in environmental and host variables such as climate and leaf senescence (Coince et al. 2014; Flessa et al. 2012). Limited cross-colonization may also further the establishment of sphere-specific communities, especially in the case of large trees (i.e. some trees were 30 m + as sampled in our study, and average height was 24 m) in which the top canopy is located at large distance from the soil, and is characterized by extreme conditions compared to the phyllosphere of shorter plants (Bodenhausen et al. 2013; Herrmann et al. 2021; Unterseher et al. 2007).
The sphere-specific adaptations of the microbial communities are exemplified by a distinction in the functions associated with the main observed taxonomic groups. The soil showed highly diverse communities (Dukunde et al. 2019; Wilhelm et al. 2023; Wubet et al. 2012), and the soil discriminant fungal taxa (e.g. Russulaceae, Elaphomycetaceae and Gloniaceae) were mainly ectomycorrhizal groups with known associations with a diversity of tree species (Castellano and Stephens 2017; Nacke et al. 2016; Spatafora et al. 2012). As indicated by our results, leaf communities showed a substantial lower bacterial alpha diversity (a factor 10 lower) and largely consisted of families which are well adapted to the phyllosphere. An example are the members of the bacterial family Hymenobacteraceae, which contain carotenoid pigments that can provide protection against the high levels of UV radiation typical for the phyllosphere (Munoz et al. 2016; Vacher et al. 2016). The identified leaf discriminant fungal taxa predominantly belonged to the very diverse fungal orders Dothideomycetes and Leotiomycetes, which are common endophytes of temperate trees (Delhomme et al. 2015; Lazarević et al. 2022; Lynikiene et al. 2020; U'Ren et al. 2012; Unterseher et al. 2013), and encompass many plant pathogens, saprobes and extremotolerant species that are associated with a broad range of hosts (Hujslová et al. 2012; Ohm et al. 2012; Zhang and Wang 2015).
Host-identity was more pronounced in the phyllosphere than the soil, both in terms of diversity and community composition. Within the soil, fungal communities showed a sharper distinction between host tree species than bacterial communities, likely related to the symbiotic association of these ectomycorrhizal fungi with specific tree species (Urbanová et al. 2015). Within the phyllosphere, the high diversity of fungi in coniferous trees is probably related to the longevity of needles compared to broadleaves, providing a more stable and longer-lived environment for the fungal community (i.e., filamentous ascomycete endophytes) to inoculate and reach a steady state (Abdelfattah et al. 2016; Flessa et al. 2012; Osono 2008). Norway spruce trees also showed a higher connectivity (number of positive network edges) and proportion of shared bacterial taxa between the bulk-soil and leaf samples, compared to European beech trees. The latter could mainly be attributed to a higher proportion of soil taxa found in the phyllosphere (Supplementary Fig. 2). Coniferous forest soils are characterised by a low pH and high concentrations of difficult to digest components such as lignin (Achilles et al. 2021; Berg 2008). Coniferous soil microbial communities are, consequentially, dominated by acidophilic taxa and saprophytic species which show resistance to environmental stressors such as drought and season (Nacke et al. 2016; Wilhelm et al. 2023), characteristics that may increase survival in the phyllosphere. Coniferous-deciduous specific differences in multi-year versus single-year leaf fall (relating to host life history traits such as leaf age) may further be an especially relevant host-driven factor in the highly seasonal temperate forests sampled here which could influence host-specific connectivity between the phyllosphere and soil. This would include (i) increasing the rate of ‘seeding’ with bacteria and fungal taxa and spores from fallen leaves on the ground during leaf senescence in deciduous forests (especially in autumn) and (ii) providing a stable environment over time for leaf communities to develop, mature and evolve in coniferous trees due to the longevity (multi-year duration) of needle leaves (Flessa et al. 2012). Other relevant differences in host attributes may include leaf surface properties (e.g. leaf morphology and area), the excretion of soluble carbohydrates and (micro) nutrients, and the production of secondary metabolites and antibiotics (Bodenhausen et al. 2014; Kembel et al. 2014; Lajoie et al. 2020). Deciduous and coniferous trees also have distinct feedback loops between trees and the surrounding soil through the formation of humus (mor, moder and mull) (Handley 1954); influencing soil fertility and properties (Nacke et al. 2016; Ponge 2013). These humus forms are characterised by different soil characteristics including pH, C:N ratio and nutrient composition, which in turn drive soil microbial community structure (Cesarz et al. 2013; Choma et al. 2020; Dukunde et al. 2019; Nacke et al. 2016). The strong link to host identity might limit the dispersal of phyllosphere communities, increasing their sensitivity to habitat fragmentation (David et al. 2016; Helander et al. 2007). This is of less concern for bulk soil communities due to their weaker link between host and microbial phylogeny and the less discrete nature of the soil environment. Other soil-associated spheres as the rhizosphere may, though, be more sensitive to habitat fragmentation due to their stronger relationship with the host (Bai et al. 2022). Overall, host identity can be considered as an important driver of microbial communities. Though less influential than the tree sphere habitat, host identity exhibits sphere-specific influences on the microbial communities impacting the fitness and evolution of the whole tree holobiont.
The characteristics of shared taxa in forest soil and phyllosphere communities
Taxa inhabiting both spheres were a major component of both the phyllosphere and soil communities in terms of relative read abundance, especially considering that only a small subset of taxa were shared between the spheres. Even though most of these taxa showed sphere-specific preferences, the capability to inhabit multiple spheres within the holobiont indicates a competitive advantage, which could be related the plant-beneficial/pathogenic capabilities of these taxa. Taxa occurring in both spheres (supplementary table 5) shared characteristics of high habitat flexibility, allowing them to adapt to the divergent environment of both the soil and top-canopy. They largely belonged to taxonomic groups with ecological phenotypic diversity and occurrence in a wide range of habitats (Bozoudi and Tsaltas 2018; Heuchert et al. 2005; Kaur et al. 2017; White et al. 1996; Willems 2014). Many of these taxa are moreover capable of N-fixation or are in other ways involved in nutrient or carbon cycling, functions that can be either part of the microbe-plant symbiosis in the rhizosphere or provide a competitive advantage in the oligotrophic phyllosphere (Azcón-Aguilar and Barea 2015; Kielak et al. 2016; King and Weber 2007; Marín and Arahal 2014). Also adaptations such as the production of pigments can be beneficial in both spheres since these can reduce competition in the soil due to their anti-bacterial properties and provide protection against UV-radiation in the canopy (Asaf et al. 2020; Pankratov and Dedysh 2010; Rashid et al. 2014). Many of these sphere-overlapping taxa show a strong association with the host. The bacterial taxa Burkholderiaceae, Conexibacter and Sphingomonas have, for example, known plant-beneficial or plant-pathogenic capabilities (some exhibit both functions based on contextual factors) (Akinola et al. 2021; Asaf et al. 2020; Romero-Gutiérrez et al. 2020). Many of the overlapping Ascomycota fungal taxa (e.g. Cladosporium and Aureobasidium pullulans), on the other hand, come from plant pathogenic families (Cooke 1959; Heuchert et al. 2005). Remarkable was the inclusion of several fruitbody forming basicomicetes (e.g. I. badia and R. cyanoxantha) in the group of overlapping taxa, which are typical arbuscular mycorrhizal soil-associated fungi (Luptáková and Mihál 2020). These taxa were possibly detected as dormant spores or carried by wind or vectors. Field observations and laboratory studies show that Blascomicota generally produce more and smaller spores than Ascomycota which can abundantly be detected in aerial samples, including samples collected above the canopy of trees (David et al. 2016; Elbert et al. 2007; Womack et al. 2015). As a more transient habitat, the phyllosphere may provide niches suitable for colonization species that easily spread (e.g., via small spores) or by r-strategists (Maignien et al. 2014). These factors can also influence the variability of phyllosphere communities, which are more sensitive to seasonal variation than soil (Coince et al. 2014; Dukunde et al. 2019; Gomes et al. 2018; Haas et al. 2018). Sampling during multiple seasons or years was, unfortunately, not possible during this study. Samples were collected during one narrow two-month window during a leaf-on season, allowing us to generate a sufficient sample number to compare host species and connectivity, and minimizing the impact of seasonal, leaf-phenological, and year-to-year alterations. Also, no distinction was made between leaf endophyte and epiphyte taxa for which there is some evidence they may form distinct communities with different environmental susceptibilities (Bodenhausen et al. 2013). Phyllosphere and soil microbial communities are shaped by a combination of specific biotic and abiotic environmental constraints, plant genotype and phenotype, and plant selective pressures (Liu et al. 2020; Llado et al. 2018). Taxa occurring in multiple spheres could provide insight in these processes and the evolution of the tree holobiont, since these taxa are strongly associated with the host tree while keeping the environmental flexibility to survive in very dissimilar environments.
Conclusions
This paper confirms the primary influence of plant sphere for microbial communities within plant holobionts, specifically in the case of tall temperate trees. While the number of taxa present across spheres was limited, they exert a disproportionate role in their communities in terms of relative abundance. These taxa generally exhibit specific ecological traits, such as ecological flexibility, growth-promotion, or pathogenic properties, which likely account for their dominance within the plant holobiont. Considering plants and their associated communities as an holobiont or “individual unit of selection” (Zilber-Rosenberg and Rosenberg 2008) may assist in linking these communities to ecosystem processes, but glazes over these differences in microbial community composition and function of the different spheres, especially between the soil and above ground components of plants. Consequentially, plant-microbiome co-evolution likely vary across spheres, in which taxa with the capability to occupy multiple spheres may play a crucial role given their evident functional connection to the host’s fitness (e.g. nutrient cycling, plant-beneficial or pathogenic traits). While this study provides a snapshot of the microbial composition at a single time point, it lays the groundwork for exploring temporal changes and deeper monitoring in subsequent research. This study highlights the need to carefully assess multiple components of the tree microbiome and their connectivity to understand and assess the ecology of temperate forests since each component tells an unique story.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Abdelfattah A, Wisniewski M, Li Destri Nicosia MG, Cacciola SO, Schena L (2016) Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PLoS One 11:e0160470. https://doi.org/10.1371/journal.pone.0160470
Achilles F, Tischer A, Bernhardt-Römermann M, Heinze M, Reinhardt F, Makeschin F, Michalzik B (2021) European beech leads to more bioactive humus forms but stronger mineral soil acidification as Norway spruce and Scots pine – Results of a repeated site assessment after 63 and 82 years of forest conversion in Central Germany. For Ecol Manage 483:118769 https://doi.org/10.1016/j.foreco.2020.118769
Akinola SA, Ayangbenro AS, Babalola OO (2021) Metagenomic insight into the community structure of maize-rhizosphere bacteria as predicted by different environmental factors and their functioning within plant proximity. Microorganisms 9:1419 https://doi.org/10.3390/microorganisms9071419
Ali AM, Darvishzadeh R, Skidmore AK, Iv D, Heiden U, Heurich M (2016) Estimating leaf functional traits by inversion of PROSPECT: assessing leaf dry matter content and specific leaf area in mixed mountainous forest. Int J Appl Earth Obs Geoinf 45:66–76. https://doi.org/10.1016/j.jag.2015.11.004
Alsos IG, Sjögren P, Edwards ME, Landvik JY, Gielly L, Forwick M, Coissac E, Brown AG, Jakobsen LV, Føreid MK, Pedersen MW (2016) Sedimentary ancient DNA from Lake Skartjørna, Svalbard: assessing the resilience of arctic flora to Holocene climate change. Holocene 26:627–642. https://doi.org/10.1177/0959683615612563
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137
Asaf S, Numan M, Khan AL, Al-Harrasi A (2020) Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit Rev Biotechnol 40:138–152. https://doi.org/10.1080/07388551.2019.1709793
Azcón-Aguilar C, Barea JM (2015) Nutrient cycling in the mycorrhizosphere. J Soil Sci Plant Nutr 15:372–396
Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:364–369. https://doi.org/10.1038/nature16192
Bai B, Liu W, Qiu X, Zhang J, Zhang J, Bai Y (2022) The root microbiome: community assembly and its contributions to plant fitness. J Integr Plant Biol 64:230–243. https://doi.org/10.1111/jipb.13226
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. https://doi.org/10.1146/annurev.arplant.57.032905.105159
Baldrian P (2017) Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol Rev 41:109–130. https://doi.org/10.1093/femsre/fuw040
Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, Hunault G, Simoneau P, Jacques M-A (2015) Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol 81:1257–1266. https://doi.org/10.1128/AEM.03722-14
Bässler C, Seifert L, Müller J (2015) The BIOKLIM project in the National Park Bavarian Forest: Lessons from a biodiversity survey. Silva Gabreta 21:81–93
Berg B (2008) Nutrient release from litter and humus in coniferous forest soils—a mini review. Scand J for Res 1:359–369. https://doi.org/10.1080/02827588609382428
Berlanga-Clavero MV, Molina-Santiago C, de Vicente A, Romero D (2020) More than words: the chemistry behind the interactions in the plant holobiont. Environ Microbiol 22:4532–4544. https://doi.org/10.1111/1462-2920.15197
Beule L, Grüning MM, Karlovsky P, lM-Arnold A (2017) Changes of scots pine phyllosphere and soil fungal communities during outbreaks of defoliating insects. Forests 8:316
Bittar TB, Pound P, Whitetree A, Moore LD, Van Stan JT (2018) Estimation of throughfall and stemflow bacterial flux in a subtropical oak-cedar forest. Geophys Res Lett 45:1410–1418. https://doi.org/10.1002/2017gl075827
Bodenhausen N, Horton MW, Bergelson J (2013) Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. https://doi.org/10.1371/journal.pone.0056329
Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA (2014) A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet 10:e1004283. https://doi.org/10.1371/journal.pgen.1004283
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vazquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Bozoudi D, Tsaltas D (2018) The multiple and versatile roles of Aureobasidium pullulans in the vitivinicultural sector. Fermentation 4:85
Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. https://doi.org/10.1146/annurev-arplant-050312-120106
Castellano MA, Stephens RB (2017) Elaphomyces species (Elaphomycetaceae, Eurotiales) from bartlett experimental forest, New Hampshire, USA. IMA Fungus 8:49–63. https://doi.org/10.5598/imafungus.2017.08.01.04
Cesarz S, Fender A-C, Beyer F, Valtanen K, Pfeiffer B, Gansert D, Hertel D, Polle A, Daniel R, Leuschner C, Scheu S (2013) Roots from beech (Fagus sylvatica L.) and ash (Fraxinus excelsior L.) differentially affect soil microorganisms and carbon dynamics. Soil Biol Biochem 61:23–32. https://doi.org/10.1016/j.soilbio.2013.02.003
Chen QL, An XL, Zhu YG, Su JQ, Gillings MR, Ye ZL, Cui L (2017) Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ Sci Technol 51:8149–8157. https://doi.org/10.1021/acs.est.7b01420
Chi F, Shen S-H, Cheng H-P, Jing Y-X, Yanni YG, Dazzo FB (2005) Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol 71:7271–7278. https://doi.org/10.1128/AEM.71.11.7271-7278.2005
Choma M, Tahovska K, Kastovska E, Barta J, Ruzek M, Oulehle F (2020) Bacteria but not fungi respond to soil acidification rapidly and consistently in both a spruce and beech forest. FEMS Microbiol Ecol 96:fiaa174. https://doi.org/10.1093/femsec/fiaa174
Coince A, Cordier T, Lengelle J, Defossez E, Vacher C, Robin C, Buee M, Marcais B (2014) Leaf and root-associated fungal assemblages do not follow similar elevational diversity patterns. PLoS One 9:e100668. https://doi.org/10.1371/journal.pone.0100668
Coluccio AE, Rodriguez RK, Kernan MJ, Neiman AM (2008) The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One 3:e2873. https://doi.org/10.1371/journal.pone.0002873
Cooke WB (1959) An ecological life history of Aureobasidium pullulans (De Bary) Arnaud. Mycopathol Mycol Appl 12:1–45. https://doi.org/10.1007/BF02118435
Cordier T, Robin C, Capdevielle X, Desprez-Loustau M-L, Vacher C (2012) Spatial variability of phyllosphere fungal assemblages: genetic distance predominates over geographic distance in a European beech stand (Fagus sylvatica). Fungal Ecol 5:509–520. https://doi.org/10.1016/j.funeco.2011.12.004
Cordier T, Barrenechea I, Lejzerowicz F, Reo E, Pawlowski J (2019) Benthic foraminiferal DNA metabarcodes significantly vary along a gradient from abyssal to hadal depths and between each side of the Kuril-Kamchatka trench. Prog Oceanogr 178:102175. https://doi.org/10.1016/j.pocean.2019.102175
David AS, Seabloom EW, May G (2016) Plant host species and geographic distance affect the structure of aboveground fungal symbiont communities, and environmental filtering affects belowground communities in a coastal dune ecosystem. Microb Ecol 71:912–926. https://doi.org/10.1007/s00248-015-0712-6
Delhomme N, Sundstrom G, Zamani N, Lantz H, Lin YC, Hvidsten TR, Hoppner MP, Jern P, Van de Peer Y, Lundeberg J, Grabherr MG, Street NR (2015) Serendipitous meta-transcriptomics: the fungal community of norway spruce (Picea abies). PLoS One 10:e0139080. https://doi.org/10.1371/journal.pone.0139080
Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF (2015) Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. PNAS 112:E1326-1332. https://doi.org/10.1073/pnas.1414261112
Dukunde A, Schneider D, Schmidt M, Veldkamp E, Daniel R (2019) Tree species shape soil bacterial community structure and function in temperate deciduous forests. Front Microbiol 10:1519. https://doi.org/10.3389/fmicb.2019.01519
Elbert W, Taylor PE, Andreae MO, Pöschl U (2007) Contribution of fungi to primary biogenic aerosols in the atmosphere: wet and dry discharged spores, carbohydrates, and inorganic ions. Atmos Chem Phys 7:4569–4588. https://doi.org/10.5194/acp-7-4569-2007
Flessa F, Peršoh D, Rambold G (2012) Annuality of Central European deciduous tree leaves delimits community development of epifoliar pigmented fungi. Fungal Ecol 5:554–561. https://doi.org/10.1016/j.funeco.2011.12.005
Fonseca-García C, Coleman-Derr D, Garrido E, Visel A, Tringe SG, Partida-Martínez LP (2016) The cacti microbiome: interplay between habitat-filtering and host-specificity. Front Microbiol 7:150. https://doi.org/10.3389/fmicb.2016.00150
Foster ZSL, Sharpton TJ, Grünwald NJ (2017) Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Comp Biol 13:e1005404. https://doi.org/10.1371/journal.pcbi.1005404
Frøslev TG, Kjøller R, Bruun HH, Ejrnæs R, Brunbjerg AK, Pietroni C, Hansen AJ (2017) Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat Commun 8:1188. https://doi.org/10.1038/s41467-017-01312-x
Gomes T, Pereira JA, Benhadi J, Lino-Neto T, Baptista P (2018) Endophytic and epiphytic phyllosphere fungal communities are shaped by different environmental factors in a mediterranean ecosystem. Microb Ecol 76:668–679. https://doi.org/10.1007/s00248-018-1161-9
Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A, Beman JM, Abell G, Philippot L, Prosser J, Foulquier A, Yuste JC, Glanville HC, Jones DL, Angel R, Salminen J, Newton RJ, Bürgmann H, Ingram LJ, Hamer U, Siljanen HMP, Peltoniemi K, Potthast K, Bañeras L, Hartmann M, Banerjee S, Yu R-Q, Nogaro G, Richter A, Koranda M, Castle SC, Goberna M, Song B, Chatterjee A, Nunes OC, Lopes AR, Cao Y, Kaisermann A, Hallin S, Strickland MS, Garcia-Pausas J, Barba J, Kang H, Isobe K, Papaspyrou S, Pastorelli R, Lagomarsino A, Lindström ES, Basiliko N, Nemergut DR (2016) Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front Microbiol 7:214. https://doi.org/10.3389/fmicb.2016.00214
Guerreiro MA, Brachmann A, Begerow D, Peršoh D (2017) Transient leaf endophytes are the most active fungi in 1-year-old beech leaf litter. Fungal Divers 89:237–251. https://doi.org/10.1007/s13225-017-0390-4
Haas JC, Street NR, Sjödin A, Lee NM, Högberg MN, Näsholm T, Hurry V (2018) Microbial community response to growing season and plant nutrient optimisation in a boreal Norway spruce forest. Soil Biol Biochem 125:197–209. https://doi.org/10.1016/j.soilbio.2018.07.005
Handley WRC (1954) Mull and mor formation in relation to forest soils. HM Stationery Office, London
Hannam KD, Quideau SA, Kishchuk BE (2007) The microbial communities of aspen and spruce forest floors are resistant to changes in litter inputs and microclimate. Appl Soil Ecol 35:635–647. https://doi.org/10.1016/j.apsoil.2006.09.005
Hassani MA, Durán P, Hacquard S (2018) Microbial interactions within the plant holobiont. Microbiome 6:58. https://doi.org/10.1186/s40168-018-0445-0
Helander M, Ahlholm J, Sieber TN, Hinneri S, Saikkonen K (2007) Fragmented environment affects birch leaf endophytes. New Phytol 175:547–553. https://doi.org/10.1111/j.1469-8137.2007.02110.x
Herrmann M, Geesink P, Richter R, Küsel K (2021) Canopy position has a stronger effect than tree species identity on phyllosphere bacterial diversity in a floodplain hardwood forest. Microb Ecol 81:157–168. https://doi.org/10.1007/s00248-020-01565-y
Heuchert B, Braun U, Schubert K (2005) Morphotaxonomic revision of fungicolous Cladosporium species (hyphomycetes). Schlechtendalia 13:1–78
Hujslová M, Kubátová A, Kostovčík M, Kolařík M (2012) Acidiella bohemica gen. et sp. nov. and Acidomyces spp. (Teratosphaeriaceae), the indigenous inhabitants of extremely acidic soils in Europe. Fungal Divers 58:33–45. https://doi.org/10.1007/s13225-012-0176-7
Kadowaki K, Sato H, Yamamoto S, Tanabe AS, Hidaka A, Toju H (2014) Detection of the horizontal spatial structure of soil fungal communities in a natural forest. Popul Ecol 56:301–310. https://doi.org/10.1007/s10144-013-0424-z
Karamanoli K, Menkissoglu-Spiroudi U, Bosabalidis AM, Vokou D, Constantinidou H-IA (2005) Bacterial colonization of the phyllosphere of nineteen plant species and antimicrobial activity of their leaf secondary metabolites against leaf associated bacteria. Chemoecology 15:59–67. https://doi.org/10.1007/s00049-005-0297-5
Kaur C, Selvakumar G, Ganeshamurthy AN (2017) Burkholderia to Paraburkholderia: the journey of a plant-beneficial-environmental bacterium. In: Shukla P (ed) Recent advances in applied microbiology. Springer, Singapore
Kembel SW, O’Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL (2014) Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. PNAS 111:13715–13720. https://doi.org/10.1073/pnas.1216057111
Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE (2016) The ecology of acidobacteria: moving beyond genes and genomes. Front Microbiol 7:744. https://doi.org/10.3389/fmicb.2016.00744
King GM, Weber CF (2007) Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5:107–118. https://doi.org/10.1038/nrmicro1595
Křenová Z, Kiener H (2013) Europe’s Wild Heart – still beating? Experiences from a new transboundary wilderness area in the middle of the Old Continent. Eur J Environ Sci 2:115–124. https://doi.org/10.14712/23361964.2015.32
Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA (2015) Sparse and compositionally robust inference of microbial ecological networks. PLoS Comp Biol 11:e1004226. https://doi.org/10.1371/journal.pcbi.1004226
Laforest-Lapointe I, Messier C, Kembel SW (2016) Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome 4:27. https://doi.org/10.1186/s40168-016-0174-1
Lajoie G, Maglione R, Kembel SW (2020) Adaptive matching between phyllosphere bacteria and their tree hosts in a neotropical forest. Microbiome 8:70. https://doi.org/10.1186/s40168-020-00844-7
Lazarević J, Topalović A, Menkis A (2022) Patterns of fungal diversity in needles, rootlets and soil of endemic Pinus peuce. Balt For 28:58. https://doi.org/10.46490/BF58
Lemanceau P, Barret M, Mazurier S, Mondy S, Pivato B, Fort T, Vacher C (2017) Chapter five - plant communication with associated microbiota in the spermosphere, rhizosphere and phyllosphere. In: Becard G (ed) Adv Bot Res. Academic Press, pp 101–133. https://doi.org/10.1016/bs.abr.2016.10.007
Levetin E, Dorsey K (2006) Contribution of leaf surface fungi to the air spora. Aerobiologia 22:3–12. https://doi.org/10.1007/s10453-005-9012-9
Lindow SE, Brandl MT (2003) Microbiology of the Phyllosphere. Appl Environ Microbiol 69:1875–1883. https://doi.org/10.1128/AEM.69.4.1875-1883.2003
Liu H, Carvalhais LC, Schenk PM, Dennis PG (2017) Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci Rep 7:41766. https://doi.org/10.1038/srep41766
Liu H, Brettell LE, Singh B (2020) Linking the phyllosphere microbiome to plant health. Trends Plant Sci 25:841–844. https://doi.org/10.1016/j.tplants.2020.06.003
Llado S, Lopez-Mondejar R, Baldrian P (2018) Drivers of microbial community structure in forest soils. Appl Microbiol Biotechnol 102:4331–4338. https://doi.org/10.1007/s00253-018-8950-4
Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL (2013) Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002. https://doi.org/10.1038/nmeth.2634
Luptáková E, Mihál I (2020) Dynamics of ectomycorrhizal mycobiota (Basidiomycota) communities on a former agricultural land (West Carpathians). Mycol Prog 19:845–857. https://doi.org/10.1007/s11557-020-01597-1
Lynikiene J, Marciulyniene D, Marciulynas A, Gedminas A, Vaiciukyne M, Menkis A (2020) Managed and unmanaged Pinus sylvestris forest stands harbour similar diversity and composition of the phyllosphere and soil fungi. Microorganisms 8:259. https://doi.org/10.3390/microorganisms8020259
MacArthur RH, Wilson EO (1967) The theory of island biogeography. Princeton University Press
Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL (2014) Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5:e00682-00613. https://doi.org/10.1128/mBio.00682-13
Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren B, Schwager EH, Chatterjee S, Thompson KN, Wilkinson JE, Subramanian A, Lu Y, Waldron L, Paulson JN, Franzosa EA, Bravo HC, Huttenhower C (2021) Multivariable association discovery in population-scale meta-omics studies. PLoS Comp Biol 17:e1009442. https://doi.org/10.1371/journal.pcbi.1009442
Marín I, Arahal DR (2014) The Family Beijerinckiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer Berlin Heidelberg, Berlin
Martins G, Lauga B, Miot-Sertier C, Mercier A, Lonvaud A, Soulas ML, Soulas G, Masneuf-Pomarede I (2013) Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS One 8:e73013. https://doi.org/10.1371/journal.pone.0073013
Munoz R, Rosselló-Móra R, Amann R (2016) Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Syst Appl Microbiol 39:281–296. https://doi.org/10.1016/j.syapm.2016.04.004
Nacke H, Goldmann K, Schöning I, Pfeiffer B, Kaiser K, Castillo-Villamizar GA, Schrumpf M, Buscot F, Daniel R, Wubet T (2016) Fine spatial scale variation of soil microbial communities under European Beech and Norway Spruce. Front Microbiol 7:2067. https://doi.org/10.3389/fmicb.2016.02067
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K (2018) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. https://doi.org/10.1093/nar/gky1022
Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, Hesse CN, Kosti I, LaButti K, Lindquist EA, Lucas S, Salamov AA, Bradshaw RE, Ciuffetti L, Hamelin RC, Kema GH, Lawrence C, Scott JA, Spatafora JW, Turgeon BG, de Wit PJ, Zhong S, Goodwin SB, Grigoriev IV (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog 8:e1003037. https://doi.org/10.1371/journal.ppat.1003037
Oksanen J, Simpson GL, Blanchet FG, Solymos P, Stevens MHH, Szoecs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, Durand S, Beatriz H, Evangelista A, Friendly M, Hannigan G, Hill MO, Lahti L, McGlinn D, Ribeiro E, Smith T, Stier A, Ter CJF, Weedon J (2022) vegan: community ecology package. R package version 2.6-4. https://CRAN.R-project.org/package=vegan
Osono T (2008) Endophytic and epiphytic phyllosphere fungi of Camellia japonica: seasonal and leaf age-dependent variations. Mycologia 100:387–391. https://doi.org/10.3852/07-110R1
Pankratov TA, Dedysh SN (2010) Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int J Syst Evol Microbiol 60:2951–2959. https://doi.org/10.1099/ijs.0.021824-0
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Peschel S, Müller CL, von Mutius E, Boulesteix A-L, Depner M (2020) NetCoMi: network construction and comparison for microbiome data in R. Briefings in Bioinformatics 22:bbaa290. https://doi.org/10.1093/bib/bbaa290
Pinheiro JC, Bates DM (2000). Mixed-Effects Models in S and S-PLUS. Springer, New York. https://doi.org/10.1007/b98882
Polling M, Sin M, de Weger LA, Speksnijder AGCL, Koenders MJF, de Boer H, Gravendeel B (2022) DNA metabarcoding using nrITS2 provides highly qualitative and quantitative results for airborne pollen monitoring. Sci Total Environ 806:150468. https://doi.org/10.1016/j.scitotenv.2021.150468
Ponge J-F (2013) Plant–soil feedbacks mediated by humus forms: a review. Soil Biol Biochem 57:1048–1060. https://doi.org/10.1016/j.soilbio.2012.07.019
Potthast K, Tischer A, Herrmann M, Weinhold A, Küsel K, van Dam NM, Michalzik B (2022) Woolly beech aphid infestation reduces soil organic carbon availability and alters phyllosphere and rhizosphere bacterial microbiomes. Plant Soil 473:639–657. https://doi.org/10.1007/s11104-022-05317-4
Pulleman M, Creamer R, Hamer U, Helder J, Pelosi C, Pérès G, Rutgers M (2012) Soil biodiversity, biological indicators and soil ecosystem services—an overview of European approaches. Curr Opin Environ Sustain 4:529–538. https://doi.org/10.1016/j.cosust.2012.10.009
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Rashid MM, Fakruddin M, Mazumdar RM, Kaniz F, Chowdhury MA (2014) Anti-bacterial activity of pigments isolated from pigment-forming soil bacteria. J Pharm Res Int 4:880–894. https://doi.org/10.9734/BJPR/2014/5148
Rodríguez-Rodríguez JC, Fenton NJ, Bergeron Y, Kembel SW (2023) Soil and tree phyllosphere microbial communities differ between coniferous and broadleaf deciduous boreal forests. Plant Soil. https://doi.org/10.1007/s11104-023-05959-y
Romero-Gutiérrez KJ, Dourado MN, Garrido LM, Olchanheski LR, Mano ET, Dini-Andreote F, Valvano MA, Araújo WL (2020) Phenotypic traits of Burkholderia spp. associated with ecological adaptation and plant-host interaction. Microbiol Res 236:126451. https://doi.org/10.1016/j.micres.2020.126451
Schneider AN, Sundh J, Sundstrom G, Richau K, Delhomme N, Grabherr M, Hurry V, Street NR (2021) Comparative fungal community analyses using metatranscriptomics and internal transcribed spacer amplicon sequencing from Norway spruce. mSystems 6:e00884-20. https://doi.org/10.1128/mSystems.00884-20
Simon J-C, Marchesi JR, Mougel C, Selosse M-A (2019) Host-microbiota interactions: from holobiont theory to analysis. Microbiome 7:5. https://doi.org/10.1186/s40168-019-0619-4
Spatafora JW, Owensby CA, Douhan GW, Boehm EW, Schoch CL (2012) Phylogenetic placement of the ectomycorrhizal genus Cenococcum in Gloniaceae (Dothideomycetes). Mycologia 104:758–765. https://doi.org/10.3852/11-233
Taberlet P, Pruhomme SM, Campione E, Roy J, Miquel C, Shehzad W, Gielly L, Rioux D, Choler P, Clément J-C, Melodelima C, Pompanon F, Coissac E (2012) Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol Ecol 21:1816–1820. https://doi.org/10.1111/j.1365-294X.2011.05317.x
Taberlet P, Bonin A, Zinger L, Coissac E (2018) Environmental DNA: for biodiversity research and monitoring. Oxford University Press, Oxford. https://doi.org/10.1093/oso/9780198767220.001.0001
Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Villarreal Ruiz L, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Poldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Partel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014) Fungal biogeography. Global diversity and geography of soil fungi. Science 346:1256688. https://doi.org/10.1126/science.1256688
Terhonen E, Blumenstein K, Kovalchuk A, Asiegbu FO (2019) Forest tree microbiomes and associated fungal endophytes: functional roles and impact on forest health. Forests 10:42
Tkacz A, Bestion E, Bo Z, Hortala M, Poole PS (2020) Influence of plant fraction, soil, and plant species on microbiota: a multikingdom comparison. mBio 11:e02785-02719. https://doi.org/10.1128/mBio.02785-19
Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM (1999) Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol 37:1846–1851. https://doi.org/10.1128/JCM.37.6.1846-1851.1999
Unterseher M, Reiher A, Finstermeier K, Otto P, Morawetz W (2007) Species richness and distribution patterns of leaf-inhabiting endophytic fungi in a temperate forest canopy. Mycol Prog 6:201–212. https://doi.org/10.1007/s11557-007-0541-1
Unterseher M, Peršoh D, Schnittler M (2013) Leaf-inhabiting endophytic fungi of European Beech (Fagus sylvatica L.) co-occur in leaf litter but are rare on decaying wood of the same host. Fungal Divers 60:43–54. https://doi.org/10.1007/s13225-013-0222-0
Urbanová M, Šnajdr J, Baldrian P (2015) Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol Biochem 84:53–64. https://doi.org/10.1016/j.soilbio.2015.02.011
U’Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE (2012) Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot 99:898–914. https://doi.org/10.3732/ajb.1100459
Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE (2016) The phyllosphere: microbial jungle at the plant-climate interface. Annu Rev Ecol Evol Syst 47:1–24. https://doi.org/10.1146/annurev-ecolsys-121415-032238
van der Knaap WO, van Leeuwen JFN, Fahse L, Szidat S, Studer T, Baumann J, Heurich M, Tinner W (2020) Vegetation and disturbance history of the Bavarian Forest National Park, Germany. Veg Hist Archaeobot 29:277–295. https://doi.org/10.1007/s00334-019-00742-5
Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytol 206:1196–1206. https://doi.org/10.1111/nph.13312
Whipps JM, Hand P, Pink D, Bending GD (2008) Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755. https://doi.org/10.1111/j.1365-2672.2008.03906.x
White DC, Sutton SD, Ringelberg DB (1996) The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol 7:301–306. https://doi.org/10.1016/S0958-1669(96)80034-6
Wilhelm RC, Muñoz-Ucros J, Weikl F, Pritsch K, Goebel M, Buckley DH, Bauerle TL (2023) The effects of mixed-species root zones on the resistance of soil bacteria and fungi to long-term experimental and natural reductions in soil moisture. Sci Total Environ 873:162266. https://doi.org/10.1016/j.scitotenv.2023.162266
Willems A (2014) The family Comamonadaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: alphaproteobacteria and betaproteobacteria. Springer, Berlin Heidelberg, Berlin
Womack AM, Artaxo PE, Ishida FY, Mueller RC, Saleska SR, Wiedemann KT, Bohannan BJM, Green JL (2015) Characterization of active and total fungal communities in the atmosphere over the Amazon rainforest. Biogeosciences 12:6337–6349. https://doi.org/10.5194/bg-12-6337-2015
Wubet T, Christ S, Schöning I, Boch S, Gawlich M, Schnabel B, Fischer M, Buscot F (2012) Differences in soil fungal communities between european beech (Fagus sylvatica L.) dominated forests are related to soil and understory vegetation. PLOS One 7:e47500. https://doi.org/10.1371/journal.pone.0047500
Yan L (2021) ggvenn: Draw venn diagram by 'ggplot2'. 0.1.9 edn
Yang H, Yang Z, Wang Q-C, Wang Y-L, Hu H-W, He J-Z, Zheng Y, Yang Y (2022) Compartment and plant identity shape tree mycobiome in a subtropical forest. Microbiol Spectr 10:e01347-e1322. https://doi.org/10.1128/spectrum.01347-22
Youngentob KN, Zdenek C, van Gorsel E (2016) A simple and effective method to collect leaves and seeds from tall trees. Methods Ecol Evol 7:1119–1123. https://doi.org/10.1111/2041-210X.12554
Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA (2015) The soil microbiome influences grapevine-associated microbiota. mBio 6:e02527-14. https://doi.org/10.1128/mBio.02527-14
Zhang N, Wang Z (2015) 3 Pezizomycotina: Sordariomycetes and Leotiomycetes. In: McLaughlin DJ, Spatafora JW (eds) Systematics and Evolution: Part B. Springer, Berlin Heidelberg, Berlin
Zhou SY, Li H, Giles M, Neilson R, Yang XR, Su JQ (2020) Microbial flow within an air-phyllosphere-soil continuum. Front Microbiol 11:615481. https://doi.org/10.3389/fmicb.2020.615481
Zhou Y, Wei Y, Zhao Z, Li J, Li H, Yang P, Tian S, Ryder M, Toh R, Yang H, Denton MD (2022) Microbial communities along the soil-root continuum are determined by root anatomical boundaries, soil properties, and root exudation. Soil Biol Biochem 171:108721. https://doi.org/10.1016/j.soilbio.2022.108721
Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. https://doi.org/10.1111/j.1574-6976.2008.00123.x
Acknowledgements
This work was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme [grant agreement number 834709]. We would like to thank M. Heurich and his team for the access to and support Bavarian Forest National Park. Our gratitude to D. Adiningrat, E. Neinavaz, T. Wang, R. Darvishzadeh, M. Lock, Y. Cheng and X. Zhu for their invaluable assistance in the field. We are also grateful to K. Zweers-Peter for her support with the DNA extractions, M. Polling for his help with the bioinformatics and H. Abdullah for checking the final manuscript. Amplicon sequencing was carried out by Genome Quebec (Montreal, Canada).
Funding
This work was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme [grant agreement number 834709].
Author information
Authors and Affiliations
Contributions
AS: Conceptualization, Methodology, Software, Investigation, Writing—Original Draft. AKS: Funding acquisition, Conceptualization, Writing—Review & Editing. AdG: Conceptualization, Writing—Review & Editing. IL: Methodology, Investigation. MR: Methodology, Investigation. YD: Methodology, Investigation. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Luz E. Bashan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Siegenthaler, A., Skidmore, A.K., de Groot, G.A. et al. Temperate tree microbiomes: divergent soil and phyllosphere microbial communities share few but dominant taxa. Plant Soil 496, 319–340 (2024). https://doi.org/10.1007/s11104-023-06364-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06364-1