Abstract
Early detection of skin cancer from skin lesion images using visual inspection can be challenging. In recent years, research in applying deep learning models to assist in the diagnosis of skin cancer has achieved impressive results. State-of-the-art techniques have shown high accuracy, sensitivity and specificity compared with dermatologists. However, the analysis of dermoscopy images with deep learning models still faces several challenges, including image segmentation, noise filtering and image capture environment inconsistency. After making the introduction to the topic, this paper firstly presents the components of machine learning-based skin cancer diagnosis. It then presents the literature review on the current advance in machine learning approaches for skin cancer classification, which covers both the traditional machine learning approaches and deep learning approaches. The paper also presents the current challenges and future directions for skin cancer classification using machine learning approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Skin cancer is a dangerous disease in Australia and worldwide, with over 5 million new cases diagnosed in the USA [1] and a high number of deaths from skin cancer in Australia each year [2]. The deadliest skin cancer is melanoma, but it is not a major form of skin cancer. Non-melanoma skin cancer, including basal cell carcinoma (BCC) and squamous cell carcinoma (SCC), make up most of the classes. In 2019, an estimated 190,000 cases of melanoma were diagnosed in the USA [3]. According to recent research, if diagnosed in the later stages, the survival rate is very poor [4]. However, the rate can go as high as 97% [5] when it is detected in the early stages, which zemphasizes the importance of the early detection of skin cancer.
In recent clinical practice, dermatologists have been zutilizing dermoscopy to diagnose skin cancer [6]. The patient’s personal data, including the history of the skin disease, exposure to sunlight, social habits and ethnicity, are also used. However, because this method usually requires a great deal of experience to identify different types of skin cancers, the diagnosis can be inconsistent among different dermatologists, and the accuracy rate can be low for inexperienced doctors [7].
Computers aiding skin cancer diagnosis methods have been well developed and have achieved great success when applied to clinical practice in the detection of dangerous skin cancer types, such as melanoma, in the last decades [8]. In 2017, Esteva et al. [9] conducted research on skin cancer classification with the HAM10000 database [10] zutilizing a pre-trained GoogleNet Inception v3 CNN model, which was proven to outperform dermatologists. Since then, a number of different deep learning (DL) models have been applied by researchers and have achieved even better accuracy on various dermoscopy datasets [11,12,13]. Although the results are promising, the techniques still face some challenges, such as variations in skin appearance, like lesion size, shape and colour, the noise of objects in the picture, and uneven lighting and contrast settings for the capture. Irregular and fuzzy borders of the skin lesion can also confuse detector programs [14, 15].
Researchers use three main types of images for the identification and classification of skin cancers: clinical images, dermoscopic images and histopathology images. In this paper, we focus on dermoscopic images, as this type is easy to retrieve and is used widely by dermatologists, and there is a good amount of public data for researchers. The International Skin Imaging Collaboration (ISIC) is the main provider of publicly accessible dermoscopy databases (Table 1). A list of databases is shown below [16, 99]:
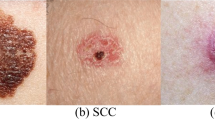
Figure 1 displays some sample images from the International Skin Imaging Collaboration 2019 dataset with, eight different types of skin cancers are shown here.
Dermoscopy is the inspection of skin lesions using a device consisting of a high-resolution lens with a proper illumination setting. Dermoscopic images are becoming a popular source for artificial intelligence (AI) studies in recent research. Given the large number of pictures in public databases, researchers have achieved impressive results.
This paper addresses the critical challenge of skin cancer, a prevalent health concern globally. Leveraging advancements in machine learning and deep learning technologies offers a promising avenue for enhancing early detection and diagnosis of skin cancers, including the lethal melanoma. Despite considerable progress, accurately diagnosing skin cancer with computational methods still presents significant obstacles, such as the variability in lesions’ appearance and the limitations of existing diagnostic tools. This paper’s objective is to comprehensively review the current landscape of machine learning applications in skin cancer diagnosis. It closely examines both traditional methodologies and the latest developments in deep learning approaches. By evaluating recent studies, their methodologies, and outcomes, this paper not only charts the progress made but also identifies persistent gaps, setting the stage for future research endeavors. This is crucial for improving the precision, reliability, and accessibility of skin cancer diagnostics, ultimately facilitating early intervention and better treatment outcomes.
Furthermore, this paper differentiates itself by thoroughly categorizing various machine learning techniques, a feature not commonly found in other reviews. While papers [96] and [98] have previously examined deep learning methods applied to skin cancer classification, our analysis extends to include subdivisions like supervised, semi-supervised, reinforcement learning, and ensemble methods. Unlike review [97], which focused solely on Convolutional Neural Networks (CNNs) within the deep learning spectrum, our examination encompasses a broader range of models, including transformers. Moreover, we explore the forefront of each specific machine learning and deep learning domain, such as K-Nearest Neighbors (KNN), Decision Trees, Support Vector Machines (SVM), and different learning models—supervised, semi-supervised, reinforcement learning, and ensemble methods. This detailed exploration of current advancements in each subfield provides researchers with a clear snapshot of the state-of-the-art, guiding their investigative pursuits. By not just listing the diverse machine learning strategies but also assessing their effectiveness and applicability in skin cancer diagnosis, this review serves as both a comprehensive overview and a directive for future research directions. It aims to inform and inspire the research community towards advancing AI-based diagnostic systems, marking the current achievements and highlighting the pathways for forthcoming innovations.
In the following sections, we explore the technologies of skin cancern detection in the last few years, starting with the background of machine learning-based skin cancer diagnosis.
2 The components of machine learning-based skin cancer diagnosis
The collection of the image, preprocessing, segmentation of the skin lesion, and classification of the lesions are the four primary processes that make up a system for machine learning-based skin cancer diagnosis. This section describes these four components.
2.1 Image collection
Training data collection is the first step in every assignment requiring machine learning. This is typically the most extensive part of the procedure. The greater the amount of data collected, the more efficient machine learning will be.
Folders or other strategies will be used to zorganize and arrange these images. In addition to converting the images to the same format, images need to be converted to grayscale and reduce their size. Each image’s size and colour format must be identical.
2.2 Preprocessing
Both traditional ML and DL techniques require input images to be preprocessed for better accuracy and more precise feature detection. Common preprocess methods for traditional techniques include greyscale, noise filtering and contrast adjustment [17]. There is also the binarisation method, which can further reduce the unwanted information contained in a grey-scaled image to achieve better algorithm performance.
2.3 Segmentation
After preprocessing, segmentation is another major process in skin lesion diagnosis. It is used to enable a classification program to focus on the diseased area instead of the healthy area.
Segmentation techniques are generally applied for disease area detection in a dermoscopy skin lesion image before conducting the classification task. This step is essential because it can reduce external noise from outside the lesion [18, 19]. In Fig. 2. One example segmented image has been displayed from the Edinburgh DERMOFIT dataset [75]. In recent years, various DL-based techniques have been developed and proven to be more effective than traditional techniques. These techniques are briefed below.
-
a.
Fully convolutional networks (FCN)
Segmentation in Edinburgh DERMOFIT dataset [75]
In 2014, FCN [26] was proposed for semantic segmentation. It first builds a downsampling (DS) path with convolutional layers and pooling layers. Adjacent to the path, there is an upsampling path containing a single layer to generate a pixel-wise prediction for segmentation.
-
b.
U-Net
The U-Net model introduced in 2015 [27] was based on the FCN. It also consists of two paths: the contraction path and the expansion path. The contraction path contains a typical convolutional network, which is made up of convolutional layers, ReLU layers and pooling layers. It is then connected to the expansion path, which combines the features and information into high-resolution feature maps through up-convolutions and concatenations. However, the expansion path contains multiple trainable layers for upsampling and uses skip connections and concatenates instead of simply adding up in the original FCN network.
-
c.
Sharp U-Net
The Sharp U-Net [95] architecture introduces an innovative approach to biomedical image segmentation, addressing the limitations of traditional U-Net by integrating depthwise convolution with a sharpening kernel filter. This method enhances the merging process of low- and high-level convolutional features, leading to clearer feature maps and reducing over- and under-segmentation issues. Unlike standard skip connections, this technique produces a sharpened feature map before combining encoder and decoder features, resulting in the fusion of semantically closer features and the reduction of artifacts early in training.
-
d.
LinkNet
Similar to U-Net, LinkNet [28] also has an encoder/contraction path connected with a decoder/expansion path. The advanced element is how the encoder is connected to the decoder. The input of each layer from the encoder is diverted to the output of the corresponding layer of the decoder, which enables the decoder to use shared spatial data from the input directly. This mechanism leads to higher efficiency due to fewer parameters used in the network.
-
e.
Other DL-based networks in recent years.
Xie et al. [29] proposed a high-resolution convolutional neural network (CNN) for skin lesion segmentation in 2020. The network contains three branches: the main high-resolution feature map, spatial attention and channel-wise attention. The results of the last two branches are fused with the output from the main branch to enhance the details of the boundaries extracted.
A hybrid method using GrabCut and the Inception v4 model for feature extraction was introduced by Sikkandar et al. in 2020 [30]. Another method combining YOLO (‘you only look once’) and GrabCut was applied [31] to skin lesion segmentation. In addition, a probabilistic-based DL model for skin lesion segmentation was developed [32] to allow an efficient mean-field approximate probabilistic inference approach to enhance a convolutional network for detecting fuzzy and irregular borders of skin lesions.
2.4 Classification
Recognizing, comprehending, and classifying concepts and things into predetermined groups or “sub-populations” is the process of classification. Machine learning systems classify upcoming datasets into categories using pre-categorized training datasets and a range of techniques. The traditional machine learning classification methods and deep learning machine learning classification methods are discussed below after the basics of machine learning are briefly presented.
2.4.1 Basics of machine learning
AI is a field of computer science in which the human brain is simulated to determine the most efficient means of achieving a given objective. Machine learning (ML) is a subfield of AI that leverages past experiences to generate future instructions without explicit programming [33]. The primary types of ML include supervised learning, unsupervised learning, semi-supervised learning and reinforcement learning.
In supervised learning, there is prior knowledge of the classes and labels for the data we are seeking to learn. There is no prior knowledge of the target classes in unsupervised learning; instead, learning is focused on zrecognizing patterns in the data. Semi-supervised learning is the combination of supervised and unsupervised learning. DL is a subsection of ML that has enhanced capabilities. The performance of both ML and DL algorithms is dependent on experience-based learning. The main difference is that DL repeats an action in order to achieve the best potential result. DL techniques take a holistic approach to the problems, whereas ML approaches follow the divide and conquer strategy. In the previous decade, significant work was conducted on improving skin cancer classification and detection through the use of both of these methodologies.
Although DL can repeat a task without human intervention, both ML and DL rely on experience to provide input. DL needs a longer training period than ML but requires less testing time. Moreover, DL requires a powerful hardware setup to work. ML performs brilliantly on low-cost hardware. ML approaches learn from prior label knowledge, while DL techniques learn from previous failures.
2.4.2 Traditional machine learning methods
Machine learning classification algorithms provide predictions about the chance that new data will fall into one of the established categories based on input training data. Recognizing, comprehending, and classifyingthings into predetermined groups or “sub-populations” is the process of classification. Machine learning systems classify upcoming datasets into categories using pre-categorized training datasets and a range of techniques. The following presents the major approaches.
K-nearest neighbour algorithms
This method compares patterns in a test set with a training set. For a vector in the target sample, the K vectors close to it in the training set are figured out, and the target sample is then assigned to the class containing the majority of the K closest neighbour vectors [20]. The classifier is simple to implement but can become time-consuming due to dataset complexity.
Decision trees
The decision tree method tries to split a dataset using given selection criteria. The dataset is repeatedly split into a tree structure, and each node of the tree represents an attribute of the dataset that is used to create decision rules for classification. The advantage of this method is it is easy to understand due to the rules it generates. The drawback is that given a complex dataset, the method may only work well with the training data [21].
Logistic regression
Logistic regression is a statistical algorithm for transforming a linear regression by a sigmoid function. It is used to compare two datasets by building decision boundaries between them and calculating the distance from the boundaries to decide the classification. The algorithm is a widely adopted method in medical image classification [22] due to its high availability in many software libraries.
Artificial neural network
An artificial neural network (ANN) is a software system designed to simulate how neurons work in the human brain to process data and information gained. An ANN contains a few artificial neurons that are structured into connected layers. Normally, an ANN needs abundant training data to feed into the network, and it can discover patterns that are difficult for humans and other program-based methodologies to find. It has become a popular research topic over the last decades [23, 24] and it is the foundation of DL.
Support vector machines
A support vector machine (SVM) is a supervised ML algorithm. It can be used for image classification problems. In this method, data items are placed in a multidimensional space, and the dimension number is the number of features in the training image. The classification is then implemented as defining hyperplanes to separate the classes and gradually find the best hyperplane. In the kernel of this method, multiple functions, such as polynomial, sigmoid and radial-based functions, can be applied to achieve the best performance [25].
2.4.3 Deep learning based classification techniques
Similar to traditional machine learning based classification algorithms, deep learning based classification algorithms provide predictions about the chance that new data will fall into one of the established categories based on input training data and current test data.This section presents various deep learning based classification techniques.
Supervised learning
A machine learning paradigm known as supervised learning is used to solve issues in which the data at hand consist of instances that have been labelled or problems wherein each data point has features and a corresponding label. Feature vectors (inputs) are converted to labels (outputs) via supervised learning algorithms, which learn a function based on sample input–output pairs. A function is inferred from the labelled training data comprising a collection of training instances [100]. Each example in supervised learning is a pair consisting of an input object (usually a vector) and a desired output value (also called the supervisory signal). An inferred function is created by a supervised learning algorithm by zanalyzing the training data and may be applied to the mapping of new cases. The method can accurately establish class labels for examples that have not been viewed in ideal situations. Based on fully labelled data, researchers have applied various methods and algorithms to improve the performance of their models. Methods like augmentation, transfer learning, network structure tuning and feature extraction algorithms have been utilized.
Supervised learning models, often require extensive computational resources for training due to the need for large labeled datasets. Training times can range from hours to weeks, depending on the complexity of the architecture and the size of the dataset. However, once trained, these models usually offer fast inference times, making them suitable for real-time applications where latency is critical. Resource usage during training is high, especially for models trained on GPUs or TPUs, but is considerably less during inference. For example, [90] shows extensive training requirements but delivers rapid inference, making it a staple in image recognition tasks.
Semi-supervised learning and self-supervised learning
Semi-supervised learning and self-supervised learning have been researched recently for skin cancer classification, as they can be suitable for scenarios in which only limited human-annotated samples are available, but a large number of unlabelled samples are accessible. By using non-annotated samples to train on pretext tasks [51] and then transferring the knowledge to a downstream task – usually supervised training using labelled data – the performance of a downstream model can be improved.
Semi-supervised learning models leverage both labelled and unlabeled data, potentially reducing the amount of labelled data required and thus training time and resource usage. These models can vary widely in their computational demands, often depending on the balance between unsupervised pre-training and supervised fine-tuning. Models like VAT (Virtual Adversarial Training) and Mean Teacher have demonstrated effectiveness in utilizing unlabeled data to reduce overall training time compared to purely supervised methods, without significantly impacting inference time [91].
Reinforcement learning
Reinforcement learning has been applied in multiple tasks, including robotics, computer games and decision support systems. Specifically, an agent is defined in the learning process, and it attempts to zmaximize the reward acquired. It can use a DL model during learning. One of the most popular deep reinforcement learning models is the deep Q-learning network (DQN) [57]. Based on the DQN, Lin et al. [58] developed an algorithm that could be used for the task of skin cancer classification. They aimed to solve imbalanced data by defining an agent that classified each image and received a reward. The reward was designed to be larger when the agent correctly classified a sample in a minority class. As a result, the agent found the optimal strategy to classify the imbalanced data.
Reinforcement learning involves learning policies based on rewards received from interactions with an environment, which can lead to varied training times depending on the complexity of the environment and the efficiency of the exploration strategy [92]. Training deep RL models is resource-intensive and can take significant time. However, the trained models can make decisions in real-time, crucial for applications like robotics and game playing.
Ensemble learning
Ensemble learning is the process of combining the outcomes of multiple DL models to achieve better overall performance. The method has been widely adopted in clustering, regression and classification tasks in the skin cancer domain. In the review of [63], a number of different approaches for building an ensemble model were introduced. The classical methods include bagging, boosting and stacking.
Bagging generally learns to use different learner models in parallel and combines the outputs through averaging/voting processes. Boosting combines different learner models in a sequential way instead of in parallel; the sequential learning strategy allows successor model learning based on the previous model. Stacking, alternatively, trains a metamodel on top of the outputs of individual models for better prediction.
The general methods for ensembles include negative correlation-based deep ensembles, explicit/implicit ensembles and homogeneous and heterogeneous ensembles [63]. Negative correlation-based deep ensembles encourage diversity among the different networks to be ensembled. Homogeneous and heterogeneous ensembles also focus on the diversity of models by zutilizing different algorithms that are not just DL-based. Explicit/implicit ensembles reduce training effort/cost by training a single model but behave like they are training an ensemble of individual models.
There are various fusion rules for combining the outputs of individual models in an ensemble [63]. These rules include the following: unweighted model averaging, majority voting, Bayes optimal classifier, stacked generalization, super learner, consensus and query by committee. Majority voting and unweighted model averaging are two of the most widely adopted methods. Some works have been conducted using majority voting as the fusion rule [70, 71]. Average output probability was used in [74]. Other rules have also been adopted, such as an ensemble weighted by cubic precision [72].
While ensemble learning can lead to better predictive performance, it also increases computational load and resource usage, both during training and inference. Techniques like boosting and bagging require training multiple models sequentially or in parallel, which can multiply the resource requirements accordingly. However, the increased accuracy and robustness of ensemble methods often justify the additional computational cost [93].
2.4.4 Advantages of traditional machine learning and deep learning
Here are some advantages that illustrate why deep learning is becoming increasingly central to the development of computer-aided diagnostic tools in dermatology and other fields of medicine.
-
a.
Automatic Feature Extraction: Deep learning models, particularly convolutional neural networks (CNNs), are capable of automatically learning features from the data without the need for manual feature extraction. This capability significantly reduces the complexity and improves the efficiency of the diagnostic process.
-
b.
Improved Diagnostic Accuracy: By employing deep learning algorithms, researchers have achieved high accuracy rates in classifying skin lesions, with some models reaching diagnostic performances comparable to that of dermatologists. This level of accuracy can greatly aid in the early detection of skin cancers, including melanoma [84].
-
c.
Efficient Use of Large Datasets: Deep learning models excel in leveraging large datasets to improve their diagnostic capabilities. The ability to train on extensive image datasets allows these models to recognize a wide variety of skin lesions, enhancing their applicability in real-world clinical settings [85].
-
d.
Handling Complex Data: Deep learning approaches are particularly well-suited for dealing with complex, high-dimensional data such as medical images. Through the use of advanced neural network architectures, deep learning models can navigate the intricacies of skin cancer images, identifying subtle patterns that may be missed by traditional methods [86].
Incorporation of Additional Data Types: Some deep learning models have been developed to combine image data with patient metadata (e.g., age, gender, skin type) to improve classification accuracy. This holistic approach mimics the diagnostic process of human experts and can lead to better outcomes [87].
However, traditional machine learning methods still have notable advantages in certain scenarios, which can be inferred from general knowledge and the context of their application before the widespread adoption of deep learning.
-
a.
Efficiency on Small Datasets: Traditional machine learning methods often require fewer data to achieve reasonable performance. In the medical field, where obtaining large annotated datasets can be challenging and expensive, these methods can still provide valuable insights [88].
-
b.
Lower Computational Requirements: Unlike deep learning models, traditional machine learning models usually have lower computational requirements for both training and inference. This makes them more accessible for use in medical institutions with limited computing resources [89].
-
c.
Flexibility in Feature Engineering: Traditional machine learning methods can benefit significantly from expertly crafted features. In situations where domain knowledge can be effectively encoded into features, these methods can outperform or complement deep learning approaches [88].
-
d.
Explainability: Traditional machine learning models, especially those that are linear or based on simple decision trees, offer a clear insight into how predictions are made. This transparency allows for easier interpretation and trust among medical practitioners, who can understand the rationale behind a model’s decision. This attribute is particularly valuable in the medical field, where the reasoning behind diagnostic decisions must be clear and justifiable [88]. While deep learning models tend to achieve higher accuracy in many tasks, their “black-box” nature makes it challenging to understand the exact reasoning behind specific decisions.
2.4.5 Evaluation criteria
Formally evaluating the performance of a classification model involves using a confusion matrix or error matrix. True positive is defined as the number of accurately zrecognized normal samples. False positive means the number of malicious samples that were misclassified. The number of normal samples that were wrongly labelled is the false negative, while the number of malicious samples that were accurately zcategorized is the true negative. These four criteria serve as the foundation for further evaluation metrics.
Commonly used evaluation metrics in skin cancer classification include accuracy, precision, sensitivity or recall, specificity and the F1 score. The definitions are provided below.
‘Accuracy’ is the ratio of the number of correctly predicted images to the total number of tested images. ‘Precision’ is the ratio of the number of correctly predicted positive images to the total number of predicted positive images of each type. ‘Sensitivity’ is the ratio of the number of correctly predicted positive images of a type to the total number of images of the type. The F1 score is the weighted average of sensitivity and precision taken into account. ‘Specificity’ is the ratio of correctly predicted negative images of a type to the total number of images that are not of the type.
3 Current advance in machine learning approaches for skin cancer classification
In recent years, researchers have employed a range of machine learning algorithms in an effort to improve the efficacy of skin cancer categorization. Following are an appraisal of some of the best performances in the research.
3.1 Traditional machine learning based technologies
This section reviews the traditional machine learning-based methods. The discussion is summarized in Table 2.
3.1.1 K-nearest neighbour algorithms
In one paper in 2022 [35], after smoothing pictures with a Gaussian filter, the authors applied an active contour model to find lesion borders and build a segmentation mask to extract lesion form attributes. They replaced the lesion pixels with those of the original picture and extracted colour and texture from the mask. Finally, they binarily classified melanoma and seborrhoeic nevi-keratosis using a K-nearest neighbour model. They achieved 0.88 accuracy on the ISIC2017 dataset for binary classification.
This paper [76] presents a MATLAB-based system that can zrecognize skin lesions and zcategorize them as normal or benign. The classification procedure is carried out by employing the K-nearest neighbour (KNN) technique to distinguish between normal skin and malignant skin lesions that indicate disease. KNN is zutilized since it is time-efficient and provides extremely precise outcomes. In terms of classifying skin lesions, the system’s accuracy achieved 98%.
3.1.2 Decision trees
[34] proposed a novel extended feature vector space technique. Colour and texture features were retrieved from images and merged to broaden the feature space. An ensemble bagged tree classifier detected melanoma using these characteristics. The experiment resulted in 0.95 accuracy, 0.94 sensitivity and 0.97 specificity on the MedNode dataset.
3.1.3 Logistic regression
The authors of [38] proposed sparse representation-based lesion image segmentation and classification. Their approach used kernel sparse representation to generate discriminative sparse codes for high-dimensional feature space representation. The discriminative kernel sparse coding formulation trained a kernel-based dictionary and linear classifier. The results showed an accuracy of 0.96, sensitivity of 0.97 and specificity of 0.93 for binary classification on the ISIC2016 dataset.
3.1.4 Support vector machines
In this paper [77], the objective of an automated system for diagnosing skin cancer from melanoma in images of skin lesions was to achieve greater accuracy with a minimum number of features. Using image processing and computer vision techniques, the researchers divided the images into lesions of interest and extracted 15 features. The system was built using a nonlinear SVM with a Gaussian radial basis function (RBF) kernel. Based on the results of the test, only six criteria were suitable for the identification of melanoma using 200 pictures. Using the optimal parameters, 86.67 percent accuracy was achieved.
Some SVM-based methods have been published recently. One paper [36] proposed a unique automated skin-melanoma detection (ASMD) system using melanoma-index. The system used picture preprocessing, BEMD, image texture improvement, entropy and energy feature mining and binary classification. Feature ranking led the system design, and Student’s t-test and other statistical approaches assessed quality. The proposed ASMD examined 600 benign and 600 DD malignant benchmark database pictures. The classification performance study showed that SVM and RBF had a classification accuracy of 0.97 on the DermIS, DermQuest and ISIC2016 datasets.
An integrated computer-aided method [37] was proposed to use recursive feature elimination-based layered structured multiclass image classification to identify each disease area. Using image processing methods, the shape, border irregularity, texture and colour of skin lesions were zanalyzed to obtain quantitative data before categorization. A layered structure classification model using an SVM classifier and radial basis function was used to test the framework’s performance. The result was an accuracy of 0.99 on the ISIC archive and an accuracy of 0.98 on the PH2 dataset.
3.2 Deep learning based technologies
This section reviews the deep learning-based methods. The discussion is summarized in Table 3.
3.2.1 Supervised learning
After using the hair-removal image-quality enhancement (HR-IQE) algorithm for preprocessing, the authors of [39] moved on to lesion segmentation using swarm intelligence algorithms to locate the region of interest (ROI), extract features from within the ROI using sped-up robust features and then narrow down the features to a select few using the grasshopper zoptimization algorithm. Finally, the pictures were classified as melanoma or non-melanoma using a custom CNN that consisted of two convolutional layers, two max-pooling layers and a flattening layer. The results showed that they achieved an accuracy of 98% on the ISIC2017 dataset and 98% on ISIC2018.
Acosta et al. [42] combined a pretrained ResNet152 structure with a mask-based and region-based CNN technique. The dataset zutilized in this study was obtained from the 2017 International Symposium on Biomedical Imaging challenge, and the findings were compared to those provided by the models used in the 2017 International Symposium on Image Computing challenge. The architecture was implemented in two stages. Mask RCNN was used to draw a boundary around the lesion, and ResNet152 was used to determine if the lesion was benign or malignant. The proposed model, eVida M6, provided a precision of 90.4 percent and a specificity of 92.5 percent, demonstrating a 3.6 percent gain in accuracy over the best-performing ISIC 2017 models.
The CNN zutilized in [40] was a custom-built one on the ISIC dataset, with five convolutional layers, five max-pooling layers, two dense layers and one dropout layer. To improve the network’s overall performance, the authors placed a significant amount of emphasis on the picture preparation effort. The experiment’s result was an F1 score of 0.96.
Sanketh et al. [78] introduced their own convolutional neural network, which consisted of two convolutional layers, two max pool layers, and a final fully connected layer. This methodology’s major purpose was to make early skin cancer diagnosis, allowing for more prompt and effective treatment and a reduced mortality rate. They used an ISIC dataset that was classed as benign or malignant. This resulted in the creation of 1,906 training shots and 816 test images. A variety of parameters had to be devised and assigned unique values before agreeing on a result of 98 percent accuracy.
A new DL model based on the VGG16 architecture was proposed in [41]. This model removed some redundant convolutional layers, added a batch znormalization (BN) layer after each pooling layer and replaced the FC layer with a global average pooling (GAP) layer. All of these changes were made to improve the model’s performance. The number of trainable parameters could be reduced by removing several convolutional layers, and the performance of the model was improved by adding BN and GAP layers without increasing the number of trainable parameters. When compared with VGG16, the network parameters may have been made smaller, allowing the overall architecture to be optimized, which in turn sped up the computation times. The result showed an accuracy of 87% on the ISIC2020 dataset.
In the paper referenced in [45], the scientists classified skin lesions using a previously trained ResNet52 network in four distinct scenarios. The following tests were carried out: training without data augmentation (DA), training with DA only on malignant images, training with DA on malignant and DS of benign images with two different proportions, and training with DA only on malignant images by including other images from different subsets within the dataset. The option that appeared to offer the optimum answer was the one in which the data were only expanded on lesions that belonged to the malignant class while at the same time keeping the ratio of malignant to benign lesions at 0.44. It achieved an accuracy of 90.4 on the ISIC2017 dataset.
In [59], the authors present a highly accurate computer-aided diagnosis approach for multi-class skin (MCS) cancer categorization. The suggested MCS cancer classification system surpassed professional dermatologists and current deep learning algorithms. They fine-tuned seven classes of HAM10000 dataset and compared five pre-trained CNNs and four ensemble models. This research reports 93.20 percent accuracy for individual models and 92.83 percent for ensemble models on ISIC2018 dataset.
In [43], the authors employed a meta-learning approach (also known as ‘learning to learn’) to automatically identify skin cancers on dermoscopic pictures on the ISIC2019 and PH2 datasets. This approach sought to understand the learning process in order to apply learned information to enhance the learning efficacy of subsequent tasks. The authors showed that the distribution of data impacted the performance of the model and that nonmedical picture attributes may be zutilized to zcategorize skin lesions. Specifically, they zutilized a ResNet50 that had already been pretrained by eliminating the last dense layer and then performing three-fold cross-validation. The results were F1 = 0.53 and Jaccard similarity index = 0.472.
The similarity measure for text processing (SMTP) is used as the loss function in the proposed system’s convolutional neural network (CNN) method [79]. They show the experimental findings obtained with various loss functions and evaluate them against the recommended SMTP loss function. In comparison to other loss functions developed for the classification issue, the proposed technique performs better. It had a 96% accuracy and a 96% specificity on PH2 dataset.
Training using preprocessed images, training with images multiplied by a segmentation mask created with U-Net and training with both of these methods were all explored in [44] using three distinct pretrained networks: EfficientNet, ResNet and SENet. The latter strategy ended up being the most precise, resulting in 88.6% accuracy on the ISIC2019 dataset.
In [45], both the VGG16 and GoogLeNet networks were put through their paces to evaluate transfer learning (TL). The optimal outcome was achieved by combining the two models, providing 81.5% accuracy on the ISIC2018 dataset.
Features were extracted from skin lesions and fed into a feedforward neural network, which used the Levenberg–Marquardt zgeneralization technique to perform classification with the goal of zminimizing the mean squared error, as described in [46]. Mean, standard deviation and skewness; entropy, mean and energy acquired by discrete 2D wavelet transform; and GLCM-obtained contrast, similarity, energy and homogeneity were all examples of the retrieved features. The result was 82.6% accuracy on the ISIC2017 dataset.
The authors of [47] proposed a multiclass multilevel classification method for multiclass (healthy, benign, malignant and eczema) classification of skin lesions and evaluated the utility of both traditional ML and cutting-edge DL techniques for this task. The first approach used a three-layered artificial neural network (ANN) to perform categorization. The previous steps of preprocessing, segmentation and feature extraction led up to that point. The second made use of TL by retraining a previously learnt AlexNet model with a dermatological dataset and making adjustments and improvements. To zmaximize efficiency, this strategy was selected. The results achieved by the DL approach were optimal at 96% on the ISIC2016 dataset.
This article [80] use deep convolutional neural networks to intelligently and rapidly classify skin cancer. This paper uses ECOC SVM and a deep convolutional neural network to classify skin cancer. Photos of RGB skin cancer obtained from the internet. Cropped photos zminimize noise. This article uses a pre-trained AlexNet convolutional neural network to extract features. Skin cancer is classified using ECOC SVM. The conclusions are based on a recommended algorithm applied to four skin cancer photographs. The implementation result reported that the mean accuracy for squamous cell carcinoma is 95.1, actinic keratosis is 98.9, and squamous cell carcinoma is 94.17 on images collected from internet.
The authors of [48] provided a skin cancer classification method that used a hybrid CNN in combination with image preprocessing. The suggested CNN had three distinct feature extraction modules. The FC layer received either a single copy of each of the blocks’ feature maps or a concatenated version of all of them. The output was mixed once everything was processed. The final experiment achieved an accuracy of 96% on the ISIC2016 dataset and an AUC of 97% on the ISIC2018 dataset.
The classification of skin lesions network is a deep CNN for multiclass skin lesion classification [49]. The network’s 68 convolutional layers are preceded by batch znormalization and LeakyRelu layers, and its basic blocks are linked. The output layer follows the GAP layer and the final FC layer. It has achieved 90% accuracy on the ISIC2019 dataset.
In [50], it was suggested that features could be added to a CNN’s layers. Specifically, dermoscopic images were segmented, and the recovered features were sent into the CNN layers as additional input. High performance in identifying different skin lesions was achieved by concatenation at the fully connected layer of both handcrafted information (such as shape, colour and texture data derived by GLCM and scatter wavelet transform) and features extracted by the CNN. The binary classification rates were 93%, 95% and 99% on the ISIC2016 dataset with different class groups.
This study [94] presents a novel framework for enhancing melanoma detection from skin lesion images through a two-stage process involving adversarial training and transfer learning. Initially, it balances the dataset by generating synthetic images of rare conditions from common ones using unpaired image-to-image translation. Then, it trains a deep convolutional neural network on this enriched dataset, employing a focal loss function to focus on harder examples. Tested on the ISIC 2016 dataset, the method achieved an AUC of 81.18% and a sensitivity of 91.76%. This approach highlights the effectiveness of combining synthetic data generation with advanced learning strategies for improved skin lesion classification.
3.2.2 Semi-supervised learning and self-supervised learning
In 2021, Ren et al. [53] proposed a method using StyleGAN as an augmentation tool and transferred an encoder of self-supervised models to a supervised learning task (downstream task) to achieve better performance on a training/test result. They first trained the StyleGAN with a non-annotated sample to generate high-resolution images. Then, a feature encoder was trained in self-supervised methods (SimCLR and BYOL) using the generated images from the previous step. Finally, a classification network was attached to the encoder for the classification job. The result showed that the accuracy of the supervised learning was improved by 1–3% after using this method on the HAM10000 [10] dataset.
Also in 2021, [52] presented a self-supervised topology clustering network for unlabelled skin cancer classification. In the method, the unlabelled skin samples were first subjected to a topology clustering algorithm for partition. Next, the data were fed into a CNN backbone network to extract low-level features. Finally, a graph network was appended for modularity maximum clustering, and a softmax layer was applied at the end to perform the classification. This method achieved an accuracy of 80.6% on unlabelled datasets..
[55] conducted an experiment to evaluate two advanced pretraining methods, SimCLR and MICLe, in self-supervised learning. They concluded that when the methods are used in pretraining, the accuracy of supervised learning with transferred knowledge can be increased by 1–2% compared with non-transferred learning.
In 2022 [54], an evaluation was conducted to compare the performance of the major self-supervised methods (pretext tasks) – BYOL, InfoMin, MoCo-V2, SimCLR and SwAV – on the skin lesion dataset ISIC 2019. The results showed that SimCLR and SwAV achieved the best performance among the five methods. [56] performed another comparison of different training pipelines of baselines, training from scratch + self-supervised learning, ImageNet pretraining + self-supervised learning and a fusion of self-supervised pretrained networks at the feature level and classification level. The fusion of self-supervised pretrained networks at the feature level turned out to be the best performer.
3.2.3 Reinforcement learning
Reinforcement learning has also been used in skin lesion segmentation recently. For example, [60] proposed a method using the Markov decision process, which addressed the issue of training an intelligent agent. Based on a set of tasks, the agent increased segmentation accuracy by applying replay memory and an action bundle as a hyperparameter. Likewise, for skin cancer image generation, a reinforcement learning model [62] was developed to assist in validating GAN-generated images to determine whether the images were real or fake.
In the work of [82], the researchers used reinforcement learning to apply nonuniform incentives and punishments based on expert-generated tables, balancing the advantages and costs of various diagnostic mistakes. When compared to supervised learning, the reinforcement learning model considerably enhanced sensitivity.
A comparison was carried out in [59] to differentiate the performance of different DL algorithms in skin cancer classification tasks, including NASNet, which is a CNN with building blocks that can be discovered and tuned using reinforcement learning. The experiment’s result showed that the NASNet model was similar in terms of performance to the Inception v3 and Xception networks.
In [61], another reinforcement method was used to enhance ANN performance by optimizing the network structure. The reinforcement Q-learning algorithm was implemented for selecting the optimal number of nodes in the hidden layer in the classification path of the network.
3.2.4 Ensemble learning
In recent years, several attempts have been made to apply various ensemble methods to skin cancer classification tasks. In the field of classical ensembles, a stacking method [64] combining the result of four fine-tuned and pretrained latest deep CNN networks was proposed, and the result showed that the ensemble model enhanced the accuracy of the skin cancer classification task by 2–3% compared with each individual model forming the ensemble. [65] also conducted an experiment on general stacking ensembles. In their work, a stacking ensemble was implemented based on a series of CNN models, and then a second ensemble combined the result of the previous ensemble and a metadata classifier. The result was also promising.
In the work of Yan et al. [66], a two-stage heterogeneous stacked ensemble model was introduced. The method first acquired the optimal feature subsets from a skin cancer dataset using a priori knowledge and a stability-based feature selection approach. The feature subsets were then zutilized to build five different models and four algorithms for stage-one stacking learning. Afterwards, a second stage of a meta-learner was appended. A metadata learner was also applied in [69] to combine with CNN classifiers to achieve better performance.
Ensemble models built at the feature level are another domain in recent research. [67] proposed an approach that extracted different feature sets, i.e. colour, shape and texture. Using different DL methods to classify different feature sets, they finally created a fusion of the methods. In [68], the top 1000 features generated by five algorithms/DL models – discrete wavelet transform, local phase quantization, local binary pattern, DarkNet19 and DarkNet53 – were chosen as the input of an SVM-based classifier. [73] proposed a method that ensembled a few different models trained on image sets. The image sets were created through a vector-level shifting algorithm. All these experiments reported good results.
Another research [81] introduced an ensemble model integrating machine learning and deep learning for skin cancer detection. This model synergizes advanced neural networks for feature extraction from images with machine learning algorithms that process these features, further refined using Contourlet Transform and Local Binary Pattern Histogram, which has achieved 93% accuracy and 99.7% recall.
The paper by Duggani et al. [83] described two new hybrid CNN models with an SVM classifier at the output layer for identifying dermoscopy pictures as benign or malignant lesions. The first and second CNN models’ retrieved features were concatenated and supplied into the SVM classifier for classification. To test the performance of the proposed model, the labels received from an expert dermatologist were employed as a reference. The experiment showed better results over the state-of-the-art CNN models.
4 Challenges and discussion
Several studies on deep learning-based skin disease diagnostics have been proposed and have shown promise in recent years with the advancement of deep learning. Before machine learning/deep learning is widely used to diagnose skin diseases in real-world clinical circumstances, there are still a number of problems that need more attention. These problems are discussed below.
-
a.
Data limitation
DL skin disease studies employ tiny image datasets for training and testing compared with image datasets for other popular DL tasks. ISIC is the largest free skin disease dataset, and tens of thousands of skin photos exist. Although it may be feasible to gather masses of data on skin disorders, without diagnoses from internet sources or hospitals, zcategorizing vast amounts of skin disease data requires professional knowledge. It is also demanding and costly. Deep neural networks require plenty of data and tagged data points. Minor tweaks increase the likelihood of overfitting the dataset. Thus, additional labelled datasets are needed for deep neural network training to identify skin problems for medical studies. Considering the challenges, more studies should focus on building DL algorithms using less skin disease diagnostic tagged data.
Furthermore, lighter-skinned people dominate skin disease databases. Dark-skinned people are equally as susceptible to skin cancer as light-skinned people, although it is usually diagnosed later. A DL system trained on lighter-skinned data may misdiagnose people of colour. In addition, current skin disease statistics exclude low-incidence categories and include only high-incidence categories. MCC, appendageal carcinoma, cutaneous lymphoma, sarcoma, Kaposi sarcoma and cutaneous secondaries are included. Consequently, if DL algorithms are trained on datasets without dark-skinned data or enough cases of rare skin diseases, they may misunderstand data on these skin conditions. It is clear that efficient skin disease detection algorithms require different skin ailment datasets.
-
b.
Image quality
Most known dermoscopic skin disease datasets are acquired using high-resolution cameras in well-lit, far-away settings. Outstanding diagnosis performance may be attained by DL algorithms trained on these high-quality skin disease datasets; nonetheless, it may be challenging for the same model to attain the same performance when evaluating photos acquired with low-resolution cameras (such as those of smartphones) under varying lighting situations and distances. DL algorithms are quite sensitive to the specifications of the camera used to take a picture. Self-captured photos typically have poor quality and considerable noise as well. Therefore, difficulties arise when using DL to diagnose skin diseases due to noise introduced by data gathered from disparate sources.
-
c.
Metadata insufficiency
In addition to evaluating the patient’s medical history, social behaviours and clinical metadata, clinicians employ ocular examinations and use medical technology (such as dermoscopy) to determine whether or not a patient has a suspected skin lesion. The histories of patients’ skin cancers, as well as their ages, sexes, ethnicities, general anatomic locations, lesion sizes and lesion structures, are important pieces of meta-diagnostic information. Occasionally, family history data is also required. It has been demonstrated that providing dermatologists, both rookies and experts, with more clinical information improves their performance. However, the vast majority of the earlier studies on using DL to diagnose skin diseases only examined photographs of the patients’ skin and not the patients’ medical histories or clinical data. A contributing factor may be that skin disease databases do not have these kinds of data.
-
d.
Frontiers
Model zgeneralization is an important topic of research in the field of skin cancer categorization. The majority of skin research has relied on medical imaging technology. When the models are zutilized with images taken with other devices, such as smartphones or conventional cameras, there is usually a considerable performance loss. Hence, TL can be useful when there is no suitable training dataset available in this new field.
Another area of research is the resilience of skin cancer classification methods. Several large training sets provide high-quality, high-resolution images; however, this has resulted in models that are less robust to noise and other sorts of disturbances. Images taken with smartphones, for example, may differ greatly in terms of illumination, angle, backdrop and quality. As a result, the model’s efficacy suffers a sudden fall.
The model’s efficacy is critical. Recent research has raised model size and training difficulty while also improving model performance. The size of a model determines whether or not it can be easily translated to multiple devices with variable processing and storage capacities. Thus, the cost of training will rise in direct proportion to the degree of difficulty involved in the training process.
The number and diversity of the training data also limit the model’s efficacy. A mismatch in the overall data quantity between the various categories is certainly a concern. Some databases provide a large amount of data for common skin cancer categories, such as BCC and SCC, whereas other less frequent but more severe skin cancer categories have little or no data. Overfitting is a phenomenon that occurs as well. To overcome this problem, samples can be created via algorithms or augmentation, and class weights in the loss function can be changed.
5 Conclusions
Despite being one of the most prevalent kinds of cancer in the world, the mortality rate associated with skin cancer is surprisingly high. The early detection and identification of skin lesions are essential for establishing the treatment that will be most beneficial to the patient and, in the event that the lesions are cancerous, for enhancing the patient’s chance of survival. Manually diagnosing this illness is a laborious process that even the most experienced dermatologists find time-consuming and demanding of their resources.
It can be difficult to detect skin cancer in its early stages based only on pictures of skin blemishes. A recent study on the use of DL models to aid in the diagnosis of skin cancer has shown amazing results. Cutting-edge approaches have provided much greater levels of accuracy, sensitivity and specificity compared with dermatologists. Nonetheless, applying DL models to zanalyze dermoscopy images has a number of challenges. These challenges include low image quality, insufficient data and inconsistency of the environment in which the image is obtained.
This paper provided an overview of many technologies that have been used to identify and classify skin cancers using dermoscopy images and particularly explored DL and traditional ML. The first stage in this approach reviewed the traditional methods for diagnosing skin cancer that do not zutilize DL. The second half of this article investigated several strategies for segmenting skin cancers. This was followed by an analysis of the most recent skin cancer classification algorithms in the five areas of supervised learning, semi-supervised learning, self-supervised learning, reinforcement learning and ensemble learning. In the last section, we discussed some of the challenges connected with using DL to classify skin cancers and the frontiers of research in the area.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Siegel RL, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62(1):10–29. https://doi.org/10.3322/caac.20138
Australian Bureau of Statistics (2019) Causes of death, Australia [internet]. Canberra: ABS. Available from: https://www.abs.gov.au/statistics/health/causes-death/causes-death-australia/2019. Accessed 1 Nov 2022
Street W (2019) Cancer facts & figures. American Cancer Society, Atlanta, GA. Available from: http://cancerstatisticscenter.cancer.org. Accessed 1 Nov 2022
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Siegel RL, Miller KD, Jemal A (2019 Jan) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–3
Vestergaard ME, Macaskill PH, Holt PE, Menzies SW (2008) Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol 159(3):669–676
Menzies SW, Bischof L, Talbot H, Gutenev A, Avramidis M, Wong L, Lo SK, Mackellar G, Skladnev V, McCarthy W, Kelly J (2005) The performance of SolarScan: an automated dermoscopy image analysis instrument for the diagnosis of primary melanoma. Arch Dermatol 141(11):1388–1396
Adeyinka AA, Viriri S (2018) Skin lesion images segmentation: a survey of the state-of-the-art. In: International conference on mining intelligence and knowledge exploration. Springer, Cham, pp 321–330
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature. 542(7639):115–118
Tschandl P, Rosendahl C, Kittler H (2018) The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data 5(1):1–9
Albahar MA (2019) Skin lesion classification using convolutional neural network with novel regularizer. IEEE Access 19(7):38306–38313
Demir A, Yilmaz F, Kose O (2019) Early detection of skin cancer using deep learning architectures: resnet-101 and inception-v3. In: 2019 medical technologies congress (TIPTEKNO). IEEE, pp 1–4
Hosny KM, Kassem MA, Foaud MM (2019) Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS One 14(5):e0217293
Al-Masni MA, Al-Antari MA, Choi MT, Han SM, Kim TS (2018) Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Prog Biomed 1(162):221–231
Naji S, Jalab HA, Kareem SA (2019) A survey on skin detection in colored images. Artif Intell Rev 15(52):1041–1087
Goyal M, Knackstedt T, Yan S, Hassanpour S (2020) Artificial intelligence-based image classification methods for diagnosis of skin cancer: challenges and opportunities. Comput Biol Med 1(127):104065
Zaqout I (2019) Diagnosis of skin lesions based on dermoscopic images using image processing techniques. Pattern Recognition-Selected Methods and Applications 15:1320
Emre Celebi M, Kingravi HA, Iyatomi H, Alp Aslandogan Y, Stoecker WV, Moss RH, Malters JM, Grichnik JM, Marghoob AA, Rabinovitz HS, Menzies SW (2008) Border detection in dermoscopy images using statistical region merging. Skin Res Technol 14(3):347–353
Emre Celebi M, Wen Q, Hwang S, Iyatomi H, Schaefer G (2013) Lesion border detection in dermoscopy images using ensembles of thresholding methods. Skin Res Technol 19(1):e252–e258
Cover T, Hart P (1967 Jan) Nearest neighbor pattern classification. IEEE Trans Inf Theory 13(1):21–27
Oates T, Jensen DD (1998) Large datasets Lead to overly complex models: an explanation and a solution. In: KDD, pp 294–298
Altman DG (2012) Practical statistics for medical research. CRC Press (1990 Nov 22)
Vennila GS, Suresh LP, Shunmuganathan KL (2012) Dermoscopic image segmentation and classification using machine learning algorithms. In: 2012 international conference on computing, electronics and electrical technologies (ICCEET). IEEE, pp 1122–1127
Freeman JA, Skapura DM (2002) Neural networks: algorithms, applications, and programming techniques. Addison Wesley Longman Publishing Co., Inc
Vapnik V (2000) The nature of statistical learning theory, 2nd edn. Springer science & business media
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany. Proceedings, part III 18 2015. Springer International Publishing, pp 234–241
Chaurasia A, Culurciello E (2017) Linknet: exploiting encoder representations for efficient semantic segmentation. In: 2017 IEEE visual communications and image processing (VCIP). IEEE, pp 1–4
Xie F, Yang J, Liu J, Jiang Z, Zheng Y, Wang Y (2020) Skin lesion segmentation using high-resolution convolutional neural network. Comput Methods Prog Biomed 186:105241
Yacin Sikkandar M, Alrasheadi BA, Prakash NB, Hemalakshmi GR, Mohanarathinam A, Shankar K (2021) Deep learning based an automated skin lesion segmentation and intelligent classification model. J Ambient Intell Humaniz Comput 12:3245–3255
Bi L, Kim J, Ahn E, Kumar A, Feng D, Fulham M (2019) Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recogn 85:78–89
Adegun AA, Viriri S, Yousaf MH (2021) A probabilistic-based deep learning model for skin lesion segmentation. Appl Sci 11(7):3025
Angra S, Ahuja S (2017) Machine learning and its applications: a review. In: 2017 international conference on big data analytics and computational intelligence (ICBDAC). IEEE, pp 57–60
Kumar S, Kumar A (2022) Extended feature space-based automatic melanoma detection system. arXiv preprint arXiv:2209.04588
Kanca E, Ayas S (2022) Learning Hand-Crafted Features for K-NN based Skin Disease Classification. In: 2022 international congress on human-computer interaction, optimization and robotic applications (HORA). IEEE, pp 1–4
Cheong KH, Tang KJ, Zhao X, Koh JE, Faust O, Gururajan R, Ciaccio EJ, Rajinikanth V, Acharya UR (2021) An automated skin melanoma detection system with melanoma-index based on entropy features. Biocybern Biomed Eng 41(3):997–1012
Chatterjee S, Dey D, Munshi S (2019 Sep) Integration of morphological preprocessing and fractal based feature extraction with recursive feature elimination for skin lesion types classification. Comput Methods Prog Biomed 1(178):201–218
Moradi N, Mahdavi-Amiri N (2019) Kernel sparse representation based model for skin lesions segmentation and classification. Comput Methods Prog Biomed 1(182):105038
Thapar P, Rakhra M, Cazzato G, Hossain MS (2022) A novel hybrid deep learning approach for skin lesion segmentation and classification. J Healthcare Eng 2022:1709842
Vanka LP, Chakravarty S (2022) Melanoma detection from skin lesions using convolution neural network. In 2022 IEEE India council international subsections conference (INDISCON). IEEE, pp. 1–5
Tabrizchi H, Parvizpour S, Razmara J (2022) An improved VGG model for skin cancer detection. Neural Process Lett 7:1–8
Jojoa Acosta MF, Caballero Tovar LY, Garcia-Zapirain MB, Percybrooks WS (2021 Dec) Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med Imaging 21(1):1–1
Garcia SI (2021) Meta-learning for skin cancer detection using deep learning techniques. arXiv preprint arXiv:2104.10775
Nadipineni H (2020) Method to classify skin lesions using dermoscopic images. arXiv preprint arXiv:2008.09418
Majtner T, Bajić B, Yildirim S, Hardeberg JY, Lindblad J, Sladoje N (2018) Ensemble of convolutional neural networks for dermoscopic images classification. arXiv preprint arXiv:1808.05071
Choudhary P, Singhai J, Yadav JS (2022 Nov) Skin lesion detection based on deep neural networks. Chemom Intell Lab Syst 15(230):104659
Hameed N, Shabut AM, Ghosh MK, Hossain MA (2020) Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert Syst Appl 1(141):112961
Hasan MK, Elahi MT, Alam MA, Jawad MT, Martí R (2022 Jan) DermoExpert: skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Inform Med Unlocked 1(28):100819
Iqbal I, Younus M, Walayat K, Kakar MU, Ma J (2021 Mar) Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput Med Imaging Graph 1(88):101843
Sekhar KS, Babu TR, Prathibha G, Vijay K, Ming LC (2021) Dermoscopic image classification using CNN with handcrafted features. Journal of king Saud University-science 33(6):101550
Jing L, Tian Y (2020) Self-supervised visual feature learning with deep neural networks: a survey. IEEE Trans Pattern Anal Mach Intell 43(11):4037–4058
Wang D, Pang N, Wang Y, Zhao H (2021 Apr) Unlabeled skin lesion classification by self-supervised topology clustering network. Biomed Signal Process Control 1(66):102428
Ren Z, Guo Y, Stella XY, Whitney D (2021) Improve image-based skin Cancer diagnosis with generative self-supervised learning. In: 2021 IEEE/ACM conference on connected health: applications, systems and engineering technologies (CHASE). IEEE, pp 23–34
Chaves L, Bissoto A, Valle E, Avila S (2021) An evaluation of self-supervised pre-training for skin-lesion analysis. arXiv preprint arXiv:210609229
Azizi S, Mustafa B, Ryan F, Beaver Z, Freyberg J, Deaton J, Loh A, Karthikesalingam A, Kornblith S, Chen T, Natarajan V (2021) Big self-supervised models advance medical image classification. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 3478–3488
Verdelho MR, Barata C (2022) On the impact of self-supervised learning in skin cancer diagnosis. In: 2022 IEEE 19th international symposium on biomedical imaging (ISBI). IEEE, pp 1–5
Tambet M (2015) Demystifying deep reinforcement learning. Computational Neuroscience Lab 2015. Available from https://neuro.cs.ut.ee/demystifying-deep-reinforcement-learning/. Accessed 1 Nov 2022
Lin E, Chen Q, Qi X (2020) Deep reinforcement learning for imbalanced classification. Appl Intell 50:2488–2502
Chaturvedi SS, Tembhurne JV, Diwan T (2020) A multi-class skin Cancer classification using deep convolutional neural networks. Multimed Tools Appl 79(39–40):28477–28498
Usmani UA, Watada J, Jaafar J, Aziz IA, Roy A (2021) A reinforcement learning algorithm for automated detection of skin lesions. Appl Sci 11(20):9367
Simin AT, Baygi SM, Noori A (2020) Cancer diagnosis based on combination of artificial neural networks and reinforcement learning. In: 2020 6th Iranian conference on signal processing and intelligent systems (ICSPIS). IEEE, pp 1–4
Annala L, Neittaanmäki N, Paoli J, Zaar O, Pölönen I (2020) Generating hyperspectral skin cancer imagery using generative adversarial neural network. In: 2020 42nd annual international conference of the IEEE engineering in Medicine & Biology Society (EMBC). IEEE, pp 1600–1603
Ganaie MA, Hu M, Malik AK, Tanveer M, Suganthan PN (2022) Ensemble deep learning: a review. Eng Appl Artif Intell 115:105151
Raza R, Zulfiqar F, Tariq S, Anwar GB, Sargano AB, Habib Z (2022) Melanoma classification from dermoscopy images using ensemble of convolutional neural networks. Mathematics 10(1):26
Lin TC, Lee HC (2020) Skin cancer dermoscopy images classification with meta data via deep learning ensemble. In: 2020 international computer symposium (ICS). IEEE, pp 237–241
Yan F, Feng Y (2022 Dec) A two-stage stacked-based heterogeneous ensemble learning for cancer survival prediction. Complex Intell Syst 8(6):4619–4639
Sabri MA, Filali Y, El Khoukhi H, Aarab A (2020) Skin cancer diagnosis using an improved ensemble machine learning model. In: 2020 international conference on intelligent systems and computer vision (ISCV). IEEE, pp 1–5
Baygin M, Tuncer T, Dogan S (2022) New pyramidal hybrid textural and deep features based automatic skin cancer classification model: ensemble DarkNet and textural feature extractor. arXiv preprint arXiv:220315090
Giovanetti A, Canalini L, Perliti SP (2022) A compact deep Ensemble for High Quality Skin Lesion Classification. InImage analysis and processing. In: ICIAP 2022 workshops: ICIAP international workshops, Lecce, Italy, May 23–27, 2022, revised selected papers, part I. Springer International Publishing, Cham, pp 510–521
Safdar K, Akbar S, Shoukat A. A majority voting based ensemble approach of deep learning classifiers for automated melanoma detection. In: 2021 international conference on innovative computing (ICIC). IEEE, pp 1–6
Kausar N, Hameed A, Sattar M, Ashraf R, Imran AS, Abidin MZ, Ali A (2021) Multiclass skin cancer classification using ensemble of fine-tuned deep learning models. Appl Sci 11(22):10593
Jiang P (2021) CNN-based diagnosis system on skin Cancer using ensemble method weighted by cubic precision. In 2021 2nd international seminar on artificial intelligence, networking and information technology (AINIT). IEEE, pp 145–1152
Thurnhofer-Hemsi K, López-Rubio E, Domínguez E, Elizondo DA (2021) Skin lesion classification by ensembles of deep convolutional networks and regularly spaced shifting. IEEE Access 9(9):112193–112205
Pratiwi RA, Nurmaini S, Rini DP, Rachmatullah MN, Darmawahyuni A (2021) Deep ensemble learning for skin lesions classification with convolutional neural network. IAES Int J Artif Intell 10(3):563
Ballerini L, Fisher RB, Aldridge B, Rees J (2013) A color and texture based hierarchical K-NN approach to the classification of non-melanoma skin lesions. In: Color medical image analysis. Springer, Dordrecht, pp 63–86
Hatem MQ (2022) Skin lesion classification system using a K-nearest neighbor algorithm. Vis Comput Ind Biomed Art 5(1):1
Mustafa S, Kimura A (2018) A SVM-based diagnosis of melanoma using only useful image features. In: 2018 international workshop on advanced image technology (IWAIT). IEEE, pp 1–4
Sanketh RS, Bala MM, Reddy PV, Kumar GP (2020) Melanoma disease detection using convolutional neural networks. In: 2020 4th international conference on intelligent computing and control systems (ICICCS). IEEE, pp 1031–1037
Patil R, Bellary S (2022) Machine learning approach in melanoma cancer stage detection. J King Saud Univ-Comput Inf Sci 34(6):3285–3293
Dorj UO, Lee KK, Choi JY, Lee M (2018) The skin cancer classification using deep convolutional neural network. Multimed Tools Appl 77:9909–9924
Tembhurne JV, Hebbar N, Patil HY, Diwan T (2023) Skin cancer detection using ensemble of machine learning and deep learning techniques. Multimed Tools Appl 16:1–24
Barata C, Rotemberg V, Codella NC, Tschandl P, Rinner C, Akay BN, Apalla Z, Argenziano G, Halpern A, Lallas A, Longo C (2023) A reinforcement learning model for AI-based decision support in skin cancer. Nat Med 29(8):1941–1946
Keerthana D, Venugopal V, Nath MK, Mishra M (2023) Hybrid convolutional neural networks with SVM classifier for classification of skin cancer. Biomed Eng Adv 1(5):100069
Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE (2018) Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol 138(7):1529–1538
Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W (2019) Deep learning to improve breast cancer detection on screening mammography. Sci Rep 9(1):12495
Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, Walliander M, Lundin M, Haglund C, Lundin J (2018) Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep 8(1):3395
Pacheco AG, Krohling RA (2021) An attention-based mechanism to combine images and metadata in deep learning models applied to skin cancer classification. IEEE J Biomed Health Inform 25(9):3554–3563
Erickson BJ, Korfiatis P, Akkus Z, Kline TL (2017) Machine learning for medical imaging radiographics. Radiographics 37(2):505–515
Ker J, Wang L, Rao J, Lim T (2017) Deep learning applications in medical image analysis. IEEE Access 29(6):9375–9389
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Miyato T, Maeda SI, Koyama M, Ishii S (2018) Virtual adversarial training: a regularization method for supervised and semi-supervised learning. IEEE Trans Pattern Anal Mach Intell 41(8):1979–1993
Schulman J, Wolski F, Dhariwal P, Radford A, Klimov O (2017) Proximal policy optimization algorithms. arXiv preprint arXiv:170706347
Cao Y, Geddes TA, Yang JY, Yang P (2020) Ensemble deep learning in bioinformatics. Nat Mach Intell 2(9):500–508
Zunair H, Hamza AB (2020) Melanoma detection using adversarial training and deep transfer learning. Phys Med Biol 65(13):135005
Zunair H, Hamza AB (2021 Sep) Sharp U-net: Depthwise convolutional network for biomedical image segmentation. Comput Biol Med 1(136):104699
Grignaffini F, Barbuto F, Piazzo L, Troiano M, Simeoni P, Mangini F, Pellacani G, Cantisani C, Frezza F (2022) Machine learning approaches for skin cancer classification from dermoscopic images: a systematic review. Algorithms 15(11):438
Bhatt H, Shah V, Shah K, Shah R, Shah M (2023) State-of-the-art machine learning techniques for melanoma skin cancer detection and classification: a comprehensive review. Intell Med 3(03):180–190
Dildar M, Akram S, Irfan M, Khan HU, Ramzan M, Mahmood AR, Alsaiari SA, Saeed AH, Alraddadi MO, Mahnashi MH (2021) Skin cancer detection: a review using deep learning techniques. Int J Environ Res Public Health 18(10):5479
Wen D, Khan SM, Xu AJ, Ibrahim H, Smith L, Caballero J, Zepeda L, de Blas PC, Denniston AK, Liu X, Matin RN (2022) Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digit Health 4(1):e64–e74
Russell SJ (2010) Artificial intelligence a modern approach. Pearson Education Inc
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Guang Yang prepared the data, conducted the review, and drafted the manuscript. Suhuai Luo reviewed the draft and enhanced the introduction, methodology, and conclusion sections. Peter Greer reviewed the manuscript and made suggestions. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not required.
Competing interests
None declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, G., Luo, S. & Greer, P. Advancements in skin cancer classification: a review of machine learning techniques in clinical image analysis. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19298-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-19298-2