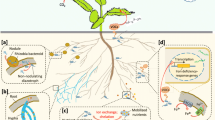
Abstract
Phosphorus (P), an essential nutrient required by plants often becomes the limiting factor for plant growth and development. Plants employ various mechanisms to sense the continuously changing P content in the soil. Transcription factors, such as SHORT ROOT (SHR), AUXIN RESPONSE FACTOR19 (ARF19), and ETHYLENE-INSENSITIVE3 (EIN3) regulate the growth of primary roots, root hairs, and lateral roots under low P. Crop improvement strategies under low P depend either on improving P acquisition efficiency or increasing P utilization. The various phosphate transporters (PTs) are involved in the uptake and transport of P from the soil to various plant cellular organelles. A plethora of regulatory elements including transcription factors, microRNAs and several proteins play a critical role in the regulation of coordinated cellular P homeostasis. Among these, the well-established P starvation signaling pathway comprising of central transcriptional factor phosphate starvation response (PHR), microRNA399 (miR399) as a long-distance signal molecule, and PHOSPHATE 2 (PHO2), an E2 ubiquitin conjugase is crucial in the regulation of phosphorus starvation responsive genes. Under PHR control, several classes of PHTs, microRNAs, and proteins modulate root architecture, and metabolic processes to enable plants to adapt to low P. Even though sucrose and inositol phosphates are known to influence the phosphorus starvation response genes, the exact mechanism of regulation is still unclear. In this review, a basic understanding of P homeostasis under low P in plants and all the above aspects are discussed.



Similar content being viewed by others
Abbreviations
- PTs:
-
Phosphate transporters
- PHT1:
-
Phosphate transporter1
- PHO2:
-
Phosphate2
- IPS1:
-
Induced by phosphate starvation1
- PHR/PSR:
-
Phosphate starvation response
- HATs:
-
High-affinity transporters
- LATs:
-
Low-affinity transporters
- MFS:
-
Major facilitator superfamily
- VPT1:
-
Vacuolar P transporter1
- sRNAs:
-
Small RNAs
- miRNA:
-
Micro RNAs
- PAP1:
-
Production of anthocyanin pigment1
- DFR:
-
Dihydroflavonol 4-reductase
- TGS:
-
Transcriptional gene silencing
- PTGS:
-
Post-transcriptional gene silencing
- PHL1:
-
PHR1 Like
- P1BS:
-
PHR1 binding site
- PHF1:
-
Phosphate transporter traffic facilitator1
- RAM:
-
Root apical meristem
- LPR1:
-
Low phosphate root1
- PDR2:
-
Phosphate deficiency response2
- SHR:
-
Short root
- EIN3:
-
Ethylene insensitive3
- RSL4:
-
Root hair defective6 like4
- TIR1:
-
Transport inhibitor response1
- ARF19:
-
Auxin response factor19
References
Ding N, Huertas R, Torres-Jerez I, Liu W, Watson B, Scheible WR, Udvardi M (2021) Transcriptional, metabolic, physiological and developmental responses of switchgrass to phosphorus limitation. Plant Cell Environ 44(1):186–202
Jiang M, Caldararu S, Zaehle S, Ellsworth DS, Medlyn BE (2019) Towards a more physiological representation of vegetation phosphorus processes in land surface models. New Phytol 222(3):1223–1229
Li Z, Hu J, Wu Y, Wang J, Song H, Chai M, Cong L, Miao F, Ma L, Tang W, Yang C (2022) Integrative analysis of the metabolome and transcriptome reveal the phosphate deficiency response pathways of alfalfa. Plant Physiol Biochem 170:49–63
Di Martino C, Crawford TW Jr (2021) 18 roles and implications of arbuscular. In: Mohammad P (ed) Handbook of plant and crop physiology, vol 12. CRC Press, Boca Raton, p 321
Etesami H, Jeong BR (2021) Contribution of arbuscular mycorrhizal fungi, phosphate-solubilizing bacteria, and silicon to P uptake by plant: a review. Front Plant Sci 12:1355
Lopez-Bucio J, De la Vega OM, Guevara-Garcia A, Herrera-Estrella L (2000) Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat Biotechnol 18(4):450–453
Gautier AT, Cochetel N, Merlin I, Hevin C, Lauvergeat V, Vivin P, Mollier A, Ollat N, Cookson SJ (2020) Scion genotypes exert long distance control over rootstock transcriptome responses to low phosphate in grafted grapevine. BMC Plant Biol 20(1):1–5
Rawat P, Das S, Shankhdhar D, Shankhdhar SC (2021) Phosphate-solubilizing microorganisms: mechanism and their role in phosphate solubilization and uptake. J Soil Sci Plant Nut 21(1):49–68
López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129(1):244–256
Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46(1):174–184
Wu M, Wei Q, Xu L, Li H, Oelmüller R, Zhang W (2018) Piriformospora indica enhances phosphorus absorption by stimulating acid phosphatase activities and organic acid accumulation in Brassica napus. Plant Soil 432(1):333–344
Brundrett MC, Tedersoo L (2018) Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol 220(4):1108–1115
Pang J, Yang J, Lambers H, Tibbett M, Siddique KH, Ryan MH (2015) Physiological and morphological adaptations of herbaceous perennial legumes allow differential access to sources of varyingly soluble phosphate. Physiol Plant 154(4):511–525
Chen A, Hu J, Sun S, Xu G (2007) Conservation and divergence of both phosphate-and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173(4):817–831
Liu F, Xu Y, Jiang H, Jiang C, Du Y, Gong C, Wang W, Zhu S, Han G, Cheng B (2016) Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int J Mol Sci 17(6):930
Liu F, Xu Y, Han G, Wang W, Li X, Cheng B (2018) Identification and functional characterization of a maize phosphate transporter induced by mycorrhiza formation. Plant Cell Physiol 59(8):1683–1694
Nagy R, Karandashov V, Chague V, Kalinkevich K, Tamasloukht MB, Xu G, Jakobsen I, Levy AA, Amrhein N, Bucher M (2005) The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J 42(2):236–250
Ezawa T, Saito K (2018) How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol 220(4):1116–1121
Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39(6):792–796
López-Bucio JS, Salmerón-Barrera GJ, Ravelo-Ortega G, Raya-González J, León P, de la Cruz HR, Campos-García J, López-Bucio J, Guevara-García ÁA (2019) Mitogen-activated protein kinase 6 integrates phosphate and iron responses for indeterminate root growth in Arabidopsis thaliana. Planta 250(4):1177–1189
Raya-González J, Ojeda-Rivera JO, Mora-Macias J, Oropeza-Aburto A, Ruiz-Herrera LF, López-Bucio J, Herrera-Estrella L (2021) MEDIATOR16 orchestrates local and systemic responses to phosphate scarcity in Arabidopsis roots. New Phytol 229(3):1278–1288
Ravelo-Ortega G, Pelagio-Flores R, López-Bucio J, Campos-García J, Reyes de la Cruz H, López-Bucio JS (2021) Early sensing of phosphate deprivation triggers the formation of extra root cap cell layers via SOMBRERO through a process antagonized by auxin signaling. Plant Mol Biol 2:1–5
Mora-Macías J, Ojeda-Rivera JO, Gutiérrez-Alanís D, Yong-Villalobos L, Oropeza-Aburto A, Raya-González J, Jiménez-Domínguez G, Chávez-Calvillo G, Rellán-Álvarez R, Herrera-Estrella L (2017) Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc Natl Acad Sci 114(17):E3563–E3572
Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon JM, Creff A, Bissler M, Brouchoud C, Hagège A, Müller J (2017) Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun 8(1):1–6
Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20(12):3258–3272
Jia H, Zhang S, Wang L, Yang Y, Zhang H, Cui H, Shao H, Xu G (2017) OsPht1; 8, a phosphate transporter, is involved in auxin and phosphate starvation response in rice. J Exp Bot 68(18):5057–5068
Hinsinger P (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237(2):173–195
Poli Y, Nallamothu V, Balakrishnan D, Ramesh P, Desiraju S, Mangrauthia SK, Voleti SR, Neelamraju S (2018) Increased catalase activity and maintenance of photosystem II distinguishes high-yield mutants from low-yield mutants of rice var. Nagina22 under low-phosphorus stress. Front Plant Sci 9:1543
Młodzińska E, Zboińska M (2016) Phosphate uptake and allocation–a closer look at Arabidopsis thaliana L. and Oryza sativa L. Front Plant Sci 7:1198
Srivastava S, Upadhyay MK, Srivastava AK, Abdelrahman M, Suprasanna P, Tran LS (2018) Cellular and subcellular phosphate transport machinery in plants. Int J Mol Sci 19(7):1914
Teng W, Zhao YY, Zhao XQ, He X, Ma WY, Deng Y, Chen XP, Tong YP (2017) Genome-wide identification, characterization, and expression analysis of PHT1 phosphate transporters in wheat. Front Plant Sci 8:543
Wan Y, Wang Z, Xia J, Shen S, Guan M, Zhu M, Qiao C, Sun F, Liang Y, Li J, Lu K (2020) Genome-wide analysis of phosphorus transporter genes in brassica and their roles in heavy metal stress tolerance. Int J Mol Sci 21(6):2209
Gho YS, Jung KH (2019) Comparative expression analyses of rice and Arabidopsis phosphate transporter families revealed their conserved roles for the phosphate starvation response. Plant Breed Biotechnol 7(1):42–49
Liu F, Chang XJ, Ye Y, Xie WB, Wu P, Lian XM (2011) Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol plant 4(6):1105–1122
Wang D, Lv S, Jiang P, Li Y (2017) Roles, regulation, and agricultural application of plant phosphate transporters. Front Plant Sci 8:817
Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, Xu G (2009) Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation. Plant J 57(5):798–809
Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1; 1 and Pht1; 4 play a major role in phosphate acquisition from both low-and high-phosphate environments. Plant J 39(4):629–642
Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24(5):2168–2183
Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31(3):341–353
Wang Y, Secco D, Poirier Y (2008) Characterization of the PHO1 gene family and the responses to phosphate deficiency of Physcomitrella patens. Plant Physiol 146(2):646–656
Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of post-translational regulation. Plant Cell 23(4):1523–1535
Ceasar SA (2020) Regulation of low phosphate stress in plants. In: Tripathi DK, Singh VP, Chauhan DK, Sharma S, Prasad SM, Dubey NK, Ramawat N (eds) Plant life under changing environment: responses and management. Academic Press, Cambridge, pp 123–156
Gu M, Chen A, Sun S, Xu G (2016) Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Mol Plant 9(3):396–416
Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216(1):23–37
Guo C, Zhao X, Liu X, Zhang L, Gu J, Li X, Lu W, Xiao K (2013) Function of wheat phosphate transporter gene TaPHT2; 1 in Pi translocation and plant growth regulation under replete and limited Pi supply conditions. Planta 237(4):1163–1178
Zhang C, Meng S, Li M, Zhao Z (2016) Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front Plant Sci 7:1398
Zhu W, Miao Q, Sun D, Yang G, Wu C, Huang J, Zheng C (2012) The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS ONE 7:e43530
Shukla V, Kaur M, Aggarwal S, Bhati KK, Kaur J, Mantri S, Pandey AK (2016) Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci Rep 6(1):1–2
Hamburger D, Rezzonico E, Petétot JM, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14(4):889–902
Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193(4):842–851
Wege S, Khan GA, Jung JY, Vogiatzaki E, Pradervand S, Aller I, Meyer AJ, Poirier Y (2016) The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol 170(1):385–400
Austin S, Mayer A (2020) Phosphate homeostasis—a vital metabolic equilibrium maintained through the INPHORS signaling pathway. Front Microbiol 11:1367
Secco D, Baumann A, Poirier Y (2010) Characterization of the rice PHO1 gene family reveals a key role for OsPHO1; 2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol 152(3):1693–1704
Salazar-Vidal MN, Acosta-Segovia E, Sanchez-Leon N, Ahern KR, Brutnell TP, Sawers RJ (2016) Characterization and transposon mutagenesis of the maize (Zea mays) Pho1 gene family. PLoS ONE 11(9):e0161882
Yamaji N, Takemoto Y, Miyaji T, Mitani-Ueno N, Yoshida KT, Ma JF (2017) Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 541(7635):92–95
Zhao H, Frank T, Tan Y, Zhou C, Jabnoune M, Arpat AB, Cui H, Huang J, He Z, Poirier Y, Engel KH (2016) Disruption of OsSULTR 3; 3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol 211(3):926–939
Tiwari M, Sharma D, Trivedi PK (2014) Artificial microRNA mediated gene silencing in plants: progress and perspectives. Plant Mol Biol 86(1–2):1–8
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151(4):2120–2132
Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53(5):731–738
Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7(3):e1002021
Huen A, Bally J, Smith P (2018) Identification and characterisation of microRNAs and their target genes in phosphate-starved Nicotiana benthamiana by small RNA deep sequencing and 5’RACE analysis. BMC Genom 19(1):1–8
Lin SI, Santi C, Jobet E, Lacut E, El Kholti N, Karlowski WM, Verdeil JL, Breitler JC, Périn C, Ko SS, Guiderdoni E (2010) Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol 51(12):2119–2131
Yan W, Chen GH, Yang LF, Gai JY, Zhu YL (2014) Overexpression of the rice phosphate transporter gene OsPT6 enhances tolerance to low phosphorus stress in vegetable soybean. Sci Hortic 177:71–76
Zhao X, Liu X, Guo C, Gu J, Xiao K (2013) Identification and characterization of microRNAs from wheat (Triticum aestivum L.) under phosphorus deprivation. J Plant Biochem Biotechnol 22(1):113–123
Valdés-López O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G (2010) MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol 187(3):805–818
Liu X, Chu S, Sun C, Xu H, Zhang J, Jiao Y, Zhang D (2020) Genome-wide identification of low phosphorus responsive microRNAs in two soybean genotypes by high-throughput sequencing. Funct Integr Genom 20(6):825–838
Gu M, Xu K, Chen A, Zhu Y, Tang G, Xu G (2010) Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol Plantarum 138(2):226–237
Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17(12):1050–1054
Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008) Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53(5):739–749
Wang F, Ding D, Li J, He L, Xu X, Zhao Y, Yan B, Li Z, Xu J (2020) Characterisation of genes involved in galactolipids and sulfolipids metabolism in maize and Arabidopsis and their differential responses to phosphate deficiency. Funct Plant Biol 47(4):279–292
Bari R, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141(3):988–999
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121(2):207–221
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15(22):2038–2043
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(16):2122–2133
Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170(1):499–514
Xue YB, Xiao BX, Zhu SN, Mo XH, Liang CY, Tian J, Liao H, Miriam G (2017) GmPHR25, a GmPHR member up-regulated by phosphate starvation, controls phosphate homeostasis in soybean. J Exp Bot 68(17):4951–4967
Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, Mao C (2015) Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168(4):1762–1776
Sega P, Pacak A (2019) Plant PHR transcription factors: put on a map. Genes 10(12):1018
Ruan W, Guo M, Wu P, Yi K (2017) Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol Biol 93(3):327–340
He Y, Zhang X, Li L, Sun Z, Li J, Chen X, Hong G (2021) SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol 230(1):205–217
Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, Shou H (2014) Rice SPX1, and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci 111(41):14953–14958
Zhang J, Jiang F, Shen Y, Zhan Q, Bai B, Chen W, Chi Y (2019) Transcriptome analysis reveals candidate genes related to phosphorus starvation tolerance in sorghum. BMC Plant Biol 19(1):1–8
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci 102(21):7760–7765
Wang H, Sun R, Cao Y, Pei W, Sun Y, Zhou H, Wu X, Zhang F, Luo L, Shen Q, Xu G (2015) OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice. Plant Cell Physiol 56(12):2381–2395
Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, Leyva A (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci 111(41):14947–14952
Osorio MB, Ng S, Berkowitz O, De Clercq I, Mao C, Shou H, Whelan J, Jost R (2019) SPX4 acts on PHR1-dependent and-independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol 181(1):332–352
Liu N, Shang W, Li C, Jia L, Wang X, Xing G, Zheng W (2018) Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol 8(1):170231
Ruan W, Guo M, Wang X, Guo Z, Xu Z, Xu L, Zhao H, Sun H, Yan C, Yi K (2019) Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol Plant 12(8):1060–1074
Gu M, Zhang J, Li H, Meng D, Li R, Dai X, Wang S, Liu W, Qu H, Xu G (2017) Maintenance of phosphate homeostasis and root development are coordinately regulated by MYB1, an R2R3-type MYB transcription factor in rice. J Exp Bot 68(13):3603–3615
Peng Z, Tian J, Luo R, Kang Y, Lu Y, Hu Y, Liu N, Zhang J, Cheng H, Niu S, Zhang J (2020) MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ 43(5):1148–1159
Chen CY, Schmidt W (2015) The paralogous R3 MYB proteins CAPRICE, TRIPTYCHON and ENHANCER OF TRY AND CPC1 play pleiotropic and partly non-redundant roles in the phosphate starvation response of Arabidopsis roots. J Exp Bot 66(15):4821–4834
Chen F, Hu Y, Vannozzi A, Wu K, Cai H, Qin Y, Mullis A, Lin Z, Zhang L (2017) The WRKY transcription factor family in model plants and crops. CRC Crit Rev Plant Sci 36(5–6):311–335
Briat JF, Rouached H, Tissot N, Gaymard F, Dubos C (2015) Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front Plant Sci 6:290
Bhalla K, Qu X, Kretschmer M, Kronstad JW (2021) The phosphate language of fungi. Trends Microbiol. https://doi.org/10.1016/j.tim.2021.08.002
Prathap V, Ali K, Singh A, Vishwakarma C, Krishnan V, Chinnusamy V, Tyagi A (2019) Starch accumulation in rice grains subjected to drought during grain filling stage. Plant Physiol Biochem 142:440–451
Liu J, Samac DA, Bucciarelli B, Allan DL, Vance CP (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41(2):257–268
Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG (2007) Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225(4):907–918
Ried MK, Wild R, Zhu J, Pipercevic J, Sturm K, Broger L, Harmel RK, Abriata LA, Hothorn LA, Fiedler D, Hiller S (2021) Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat Commun 12(1):1–3
Azevedo C, Saiardi A (2017) Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem Sci 42(3):219–231
Chien PS, Chiang CP, Leong SJ, Chiou TJ (2018) Sensing and signaling of phosphate starvation: from local to long distance. Plant Cell Physiol 59(9):1714–1722
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
PV wrote the first draft of the review article. Whereas AT and AK have contributed equally to the literature collection, and manuscript documentation. CM and all authors contributed to the revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Prathap, V., Kumar, A., Maheshwari, C. et al. Phosphorus homeostasis: acquisition, sensing, and long-distance signaling in plants. Mol Biol Rep 49, 8071–8086 (2022). https://doi.org/10.1007/s11033-022-07354-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07354-9