Abstract
Key message
OsPHR4 mediates the regulation of Pi-starvation signaling and Pi-homeostasis in a PHR1-subfamily dependent manner in rice.
Abstract
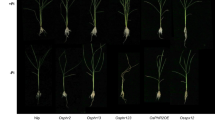
Phosphate (Pi) starvation response is a sophisticated process for plant in the natural environment. In this process, PHOSPHATE STARVATION RESPONSE 1 (PHR1) subfamily genes play a central role in regulating Pi-starvation signaling and Pi-homeostasis. Besides the three PHR1 orthologs in Oryza sativa L. (Os) [(Os) PHR1, (Os) PHR2, and (Os) PHR3], which were reported to regulated Pi-starvation signaling and Pi-homeostasis redundantly, a close related PHR1 ortholog [designated as (Os) PHR4] is presented in rice genome with unknown function. In this study, we found that OsPHR4 is a Pi-starvation induced gene and mainly expresses in vascular tissues through all growth and development periods. The expression of OsPHR4 is positively regulated by OsPHR1, OsPHR2 and OsPHR3. The nuclear located OsPHR4 can respectively interact with other three PHR1 subfamily members to regulate downstream Pi-starvation induced genes. Consistent with the positive role of PHR4 in regulating Pi-starvation signaling, the OsPHR4 overexpressors display higher Pi accumulation in the shoot and elevated expression of Pi-starvation induced genes under Pi-sufficient condition. Besides, moderate growth retardation and repression of the Pi-starvation signaling in the OsPHR4 RNA interfering (RNAi) transgenic lines can be observed under Pi-deficient condition. Together, we propose that OsPHR4 mediates the regulation of Pi-starvation signaling and Pi-homeostasis in a PHR1-subfamily dependent manner in rice.








Similar content being viewed by others

References
Ai PH, Sun SB, Zhao JN, Fan XR, Xin WJ, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, Xu GH (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57:798–809
Bournier M, Tissot N, Mari S, Boucherez J, Lacombe E, Briat JF, Gaymard F (2013) Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J Biol Chem 288:22670–22680
Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6:e1001102
Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59:2479–2497
Calvenzani V, Testoni B, Gusmaroli G, Lorenzo M, Gnesutta N, PetroniK, Mantovani R, Tonelli C (2012) Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7:e42902
Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia QJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36:105–113
Cheng L, Bucciarelli B, Liu J, Zinn K, Miller S, Patton-Vogt J, Allan D, Shen J, Vance CP (2011) White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol 156:1131–1148
Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62:185–206
Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2:43–58
Guo MN, Ruan WY, Li CY, Huang FL, Zeng M, Liu YY, Yu YN, Ding XM, Wu YR, Wu ZC, Mao CZ, Yi KK, Wu P, Mo XR (2015) Integrative comparison of the role of the PHOSPHATE STARVATION RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168:1762–1776
Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132:578–596
Hernandez G, Ramirez M, Valdes-Lopez O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, Wu HC, Lara M, Town CD, Kopka J, Udvardi MK, Vance CP (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144:752–767
Hinsinger P, Betencourt E, Bernard L, Brauman A, Plassard C, Shen J, Tang X, Zhang F (2011) P for two, sharing a scarce resource: soilphosphorus acquisition in the rhizosphere of intercropped species. Plant Physiol 156:1078–1086
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions:β-glucuronidaseas a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Khan GA, Bouraine S, Wege S, Li YY, de Carbonnel M, Berthomieu P, Poirier Y, Rouached H (2014) Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J Exp Bot 65:871–884
Liu F, Wang ZY, Ren HY, Shen CJ, Li Y, Ling HQ, Wu CY, Lian XM, Wu P (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62:508–517
Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrixgene chips determined plant responses to phosphate deprivation. Proc Natl Acad of Sci USA 102:11934–11939
Morcuende R, Bari R, Gibon Y, Zheng WM, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible WR (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30:85–112
Nilsson L, Lundmark M, Jensen PE, Nielsen TH (2012) The Arabidopsis transcription factor PHR1 is essential for adaptation to high light and retaining functional photosynthesis during phosphate starvation. Physiol Plant 144:35–47
Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2:83
O’Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu JQ, Rydeen A, Bozsoki Z, Uhde-Stone C, Tu ZJ, Allan D, Gronwald JW, Vance CP (2013) An RNA-seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161:705–724
Pant BD, Burgos A, Pant P, Cuadros-Inostroza A, Willmitzer L, Scheible WR (2015a) The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J Exp Bot 66:1907–1918
Pant BD, Pant P, Erban A, Huhman D, Kopka J, Scheible WR (2015b) Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ 38:172–187
Raghothama KG (2000) Phosphate transport and signaling. Curr Opin in Plant Biol 3:182–187
Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS One 7:e44005
Richardson AE, Hocking PJ, Simpson RJ, George TS (2009) Plant mechanisms to optimize access to soil phosphorus. Crop Pasture Sci 60:124–143
Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3:288–299
Rouached H, Secco D, Arpat B, Poirier Y (2011) The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol 11(1):19
Ruan W, Guo M, Cai L, Hu H, Li C, Liu Y, Wu Z, Mao C, Yi K, Wu P, Mo X (2015) Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol Biol 87:429–440
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Secco D, Jabnoune M, Walker H, Shou HX, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice rootsand shoots in response to phosphate starvation and recovery. Plant Cell 25:4285–4304
Sun LC, Song L, Zhang Y, Zheng Z, Liu D (2015) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170:499–514
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157:423–447
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12
Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F et al (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signaling and increases grain yield in wheat. AnnBot (Lond) 111:1139–1153
Wu P, Ma LG, Hou XL, Wang MY, Wu YR, Liu FY, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132:1260–1271
Wu ZC, Ren HY, McGrath SP, Wu P, Zhao FJ (2011) Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol 157:498–508
Wu P, Shou HX, Xu GH, Lian XM (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 16:1–8
Yamaji N, Ma JF (2014) The node, a hub for nutrient distribution in gramineous plants. Trends Plant Sci 19:556–563
Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146:1673–1686
Acknowledgements
This research was supported by the Ministry of Science and Technology Key R&D program (2016YFD0100700) and the National Natural Science Foundation of China (31601807, 31272227, 31322048).
Author contributions
KKY, PW and WYR designed the research. WYR and MNG performed the experiments. KKY and WYR wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wenyuan Ruan and Meina Guo contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ruan, W., Guo, M., Wu, P. et al. Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol Biol 93, 327–340 (2017). https://doi.org/10.1007/s11103-016-0564-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0564-6