Abstract
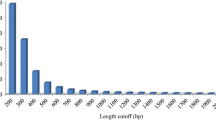
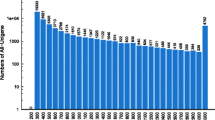
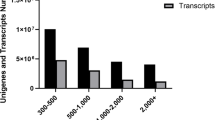
Lateolabrax japonicus is an abundant marine aquatic fish species that is commonly cultured in East Asia due to its high commercial value. In this study, RNA-Seq analysis of L. japonicus was carried out to identify reproduction- and growth-related genes expressed in L. japonicus ovaries at different ages using Illumina sequencing technology. In total, 334,388,688 high-quality reads were obtained in four libraries, i.e., 4-year-old ovaries (4th_Ovary), 3-year-old ovaries (3rd_Ovary), 2-year-old ovaries (2nd_Ovary), and 1-year-old ovaries (1st_Ovary). The reads were then de novo assembled into 101,860 unigenes with an average unigene length of 879 bp. In total, 30,142 unigenes (29.59%) were annotated in public databases, including Nr database (Nr), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Cluster of Orthologous Groups of proteins (COG), based on sequence similarity. Comparative analysis revealed that there were 35,749, 43,657, and 36,819 differentially expressed genes (DEGs) in three comparisons (4th_Ovary versus 3rd_Ovary, 4th_Ovary versus 2rd_Ovary, and 4th_Ovary versus 1st_Ovary, respectively). In total, 24,295 DEGs were different expressed in 4th_Ovary. Enrichment and pathway analyses of the DEGs were also carried out to excavate the candidate genes related to reproduction and growth, and 402 genes that potential involved in the regulation of reproduction and growth were identified, e.g., GnRHR (GnRH receptor), GHR 2 (growth hormone receptor 2), I_LGF1R (insulin-like growth factor 1 receptor), etc. Our findings expanded the genomic resources of L. japonicus and provided fundamental information for further studies.











Similar content being viewed by others
References
Schuler G, Boguski M, Stewart E, Stein L, Gyapay G, Rice K, White R, Rodriguez-Tome P, Aggarwal A, Bajorek E (1996) A gene map of the human genome. Science 274(5287):540–546
Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O’Connor TJ (1996) A comprehensive genetic map of the mouse genome. Nature 380(6570):149–152
Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L (2010) The genome of the Western clawed frog Xenopus tropicalis. Science 328(5978):633–636
Groenen MA, Cheng HH, Bumstead N, Benkel BF, Briles WE, Burke T, Burt DW, Crittenden LB, Dodgson J, Hillel J (2000) A consensus linkage map of the chicken genome. Genome Res 10(1):137–147
Postlethwait JH, Yan Y-L, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z (1998) Vertebrate genome evolution and the zebrafish gene map. Nat Genet 18(4):345–349
Wang W, Wu X, Liu Z, Zheng H, Cheng Y (2014) Insights into hepatopancreatic functions for nutrition metabolism and ovarian development in the crab Portunus trituberculatus: gene discovery in the comparative transcriptome of different hepatopancreas stages. PLoS ONE 9(1):e84921
Kocher TD (2004) Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet 5(4):288–298
Venkatesh B (2003) Evolution and diversity of fish genomes. Curr Opin Genet Dev 13(6):588–592
Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y (2007) The medaka draft genome and insights into vertebrate genome evolution. Nature 447(7145):714–719
Xiang L-x, He D, Dong W-r, Zhang Y-w, Shao J-z (2010) Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics 11(1):472
Hegedűs Z, Zakrzewska A, Ágoston VC, Ordas A, Rácz P, Mink M, Spaink HP, Meijer AH (2009) Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol 46(15):2918–2930
Stockhammer OW, Zakrzewska A, Hegedûs Z, Spaink HP, Meijer AH (2009) Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol 182(9):5641–5653
Xie Z, Xiao l, Wang D, Fang C, Liu Q, Li Z, Liu X, Zhang Y, Li S, Lin H (2014) Transcriptome analysis of the Trachinotus ovatus: identification of reproduction, growth and immune-related genes and microsatellite markers. PLos ONE 9(10):e109419
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18(1):53–63
Gjerde B (1986) Growth and reproduction in fish and shellfish. Aquaculture 57(1):37–55
Sarà G, Reid G, Rinaldi A, Palmeri V, Troell M, Kooijman S (2012) Growth and reproductive simulation of candidate shellfish species at fish cages in the Southern Mediterranean: Dynamic Energy Budget (DEB) modelling for integrated multi-trophic aquaculture. Aquaculture 324:259–266
Le Gac F, Blaise O, Fostier A, Le Bail P-Y, Loir M, Mourot B, Weil C (1993) Growth hormone (GH) and reproduction: a review. Fish Physiol Biochem 11(1–6):219–232
Donelson J, Munday P, McCormick M, Pitcher C (2012) Rapid transgenerational acclimation of a tropical reef fish to climate change. Nat Clim Chang 2(1):30–32
Runnalls TJ, Beresford N, Losty E, Scott AP, Sumpter JP (2013) Several synthetic progestins with different potencies adversely affect reproduction of fish. Environ Sci Technol 47(4):2077–2084
Schreck CB, Contreras-Sanchez W, Fitzpatrick MS (2001) Effects of stress on fish reproduction, gamete quality, and progeny. Aquaculture 197(1):3–24
Zhenzhen X, Ling X, Dengdong W, Chao F, Qiongyu L, Zihao L, Xiaochun L, Yong Z, Shuisheng L, Haoran L (2014) Transcriptome analysis of the Trachinotus ovatus: identification of reproduction, growth and immune-related genes and microsatellite markers. PLoS ONE 9(10):e109419
Wang W, Yi Q, Ma L, Zhou X, Zhao H, Wang X, Qi J, Yu H, Wang Z, Zhang Q (2014) Sequencing and characterization of the transcriptome of half-smooth tongue sole (Cynoglossus semilaevis). BMC Genomics 15(1):470
Reading BJ, Chapman RW, Schaff JE, Scholl EH, Opperman CH, Sullivan CV (2012) An ovary transcriptome for all maturational stages of the striped bass (Morone saxatilis), a highly advanced perciform fish. BMC Res Notes 5(1):111
Men K, Ai Q, Mai K, Xu W, Zhang Y, Zhou H (2014) Effects of dietary corn gluten meal on growth, digestion and protein metabolism in relation to IGF-I gene expression of Japanese seabass, Lateolabrax japonicus. Aquaculture 428:303–309
Wang J, Ai Q, Mai K, Xu H, Zuo R, Xu W, Zhang W, Zhang C (2014) Effects of dietary ethoxyquin on growth, feed utilization and residue in the muscle of juvenile Japanese seabass, Lateolabrax japonicus. Aquac Res 46(11):2656–2664
Li Y, Ai Q, Mai K, Xu W, Deng J, Cheng Z (2014) Comparison of high-protein soybean meal and commercial soybean meal partly replacing fish meal on the activities of digestive enzymes and aminotransferases in juvenile Japanese seabass, Lateolabrax japonicus (Cuvier, 1828). Aquac Res 45(6):1051–1060
Jia P, Jia K-T, Yi M-S (2015) Complete genome sequence of a fish nervous necrosis virus isolated from sea perch (Lateolabrax japonicus) in China. Genome Announc 3(3):e00048–e00015
Xue M, Luo L, Wu X, Ren Z, Gao P, Yu Y, Pearl G (2006) Effects of six alternative lipid sources on growth and tissue fatty acid composition in Japanese sea bass (Lateolabrax japonicus). Aquaculture 260(1):206–214
Wang Y, Cong B, Shen J, Liu S, Liu F, Wang N, Huang X (2012) Molecular cloning and functional analysis of a voltage-gated potassium channel in lymphocytes from sea perch, Lateolabrax japonicus. Fish Shellfish Immunol 33(3):605–613
C. Zhang, M. Li (2005) Advance of study on reproductive biology and breeding technology of Lateolabrax aponicus. J Ningbo Univ (Nat Sci Eng Edn) 3:032
Kang DY, Han HK, Baek HJ (2002) Monthly gonadal and sex hormonal changes of indoor-reared seabass, Lateolabrax japonicus during annual reproductive cycle. Korean J Fish Aquat Sci 35(6):614–620
Hayashi I (1972) On the ovarian maturation of the Japanese sea bass, Lateolabrax japonicus. Jpn J Ichthyol 19(4):243–254
Xu J, Yan B, Teng Y, Lou G, Lu Z (2010) Analysis of nutrient composition and fatty acid profiles of Japanese sea bass Lateolabrax japonicus (Cuvier) reared in seawater and freshwater. J Food Compos Anal 23(5):401–405
Xu B, Zhang P, Miao H, Li D, Moriyama S, Kawauchi H (1997) Isolation and bioactivity of growth hormone from Japanese seabass Lateolabrax japonicus. Acta Zool Sin 44(2):170–178
Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12(10):1611–1618
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31(1):365–370
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN (2003) The COG database: an updated version includes eukaryotes. BMC Bioinform 4(1):41
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34(suppl 2):W293–W297
Audic S, Claverie J-M (1997) The significance of digital gene expression profiles. Genome Res 7(10):986–995
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95(25):14863–14868
Saldanha AJ (2004) Java Treeview—extensible visualization of microarray data. Bioinformatics 20(17):3246–3248
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25(4):402–408
Long Y, Li Q, Zhou B, Song G, Li T, Cui Z (2013) De novo assembly of mud loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion. PLoS ONE 8(2):e56998
Wu J, Stoica BA, Dinizo M, Pajoohesh-Ganji A, Piao C, Faden AI (2012) Delayed cell cycle pathway modulation facilitates recovery after spinal cord injury. Cell Cycle 11(9):1782–1795
Du Y-X, Ma K-Y, Qiu G-F (2015) Discovery of the genes in putative GnRH signaling pathway with focus on characterization of GnRH-like receptor transcripts in the brain and ovary of the oriental river prawn Macrobrachium nipponense. Aquaculture 442:1–11
Shi Y, He M (2014) Differential gene expression identified by RNA-Seq and qPCR in two sizes of pearl oyster (Pinctada fucata). Gene 538(2):313–322
Ge G, Xiao P, Zhang Y, Yang L (2011) The first insight into the tissue specific taxus transcriptome via Illumina second generation sequencing. PLoS ONE 6(6):e21220
Salem M, Rexroad CE, Wang J, Thorgaard GH, Yao J (2010) Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11(1):564
Mair GC, Abucay JS, Beardmore JA, Skibinski DO (1995) Growth performance trials of genetically male tilapia (GMT) derived from YY-males in Oreochromis niloticus L.: on station comparisons with mixed sex and sex reversed male populations. Aquaculture 137(1):313–323
Palmieri JR (1977) Host-induced morphological variations in the strigeoid trematode posthodiplostomum minimum (trematoda: diplostomatidae). ii. body measurements and tegument modifications. Gt Basin Nat 37(2):129–137
Guraya SS, Kaur R, Saxena PK (1975) Morphology of ovarian changes during the reproductive cycle of the fish Mystus tengara (Ham.). Cells Tissues Organs 91(2):222–260
Schmid AC, Näf E, Kloas W, Reinecke M (1999) Insulin-like growth factor-I and-II in the ovary of a bony fish, Oreochromis mossambicus, the tilapia: in situ hybridisation, immunohistochemical localisation, Northern blot and cDNA sequences. Mol Cell Endocrinol 156(1):141–149
Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC (2005) Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Can Res 65(1):338–348
Zhou P, Liu DJ, Cang M, Ma YZ, Yang DS, Li HJ, Wang LM, Bou S, Feng HL (2008) TGFα and EGFR in ovine preimplantation embryos and effects on development. Anim Reprod Sci 104(2–4):370–381
Gothilf Y, Elizur A, Chow M, Chen TT, Zohar Y (1995) Molecular cloning and characterization of a novel gonadotropin-releasing hormone from the gilthead seabream (Sparus aurata). Mol Mar Biol Biotechnol 4(1):27
Jansen HT, Cutter C, Hardy S, Lehman MN, Goodman RL (2003) Seasonal plasticity within the gonadotropin-releasing hormone (GnRH) system of the ewe: changes in identified GnRH inputs and glial association. Endocrinology 144(8):3663–3676
Kuiper GG, Enmark E, Peltohuikko M, Nilsson S, Gustafsson JA (1996) Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA 93(12):5925
Leung YK, Mak P, Hassan S, Ho SM (2006) Estrogen receptor (ER)-β isoforms: a key to understanding ER-β signaling. Proc Natl Acad Sci USA 103(35):13162–13167
Kitano T, Koyanagi T, Adachi R, Sakimura N, Takamune K, Abe SI (2006) Assessment of estrogenic chemicals using an estrogen receptor α (ERα)- and ERβ-mediated reporter gene assay in fish. Mar Biol 149(1):49–55
Acknowledgements
This work was supported by The Central Public-Interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2017YB03, 2017YB28 and 2018ZD01); the Natural Science Foundation of China (3180228), the Natural Science Foundation of Guangdong Province, China (c18140500000602).
Author information
Authors and Affiliations
Contributions
LHQ initiated the project. CZ performed bioinformatics analyses, designed experiments, carried out experiments, analyzed the data and interpreted results and wrote the manuscript. PFW reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2018_4383_MOESM2_ESM.xlsx
Additional File 2. The functional classification of the COG classes for the transcriptome of Lateolabrax japonicus. (XLSX 122 KB)
11033_2018_4383_MOESM3_ESM.xlsx
Additional File 3. The functional classification of the GO classes for the transcriptome of Lateolabrax japonicus. (XLSX 309 KB)
11033_2018_4383_MOESM5_ESM.xlsx
Additional File 5. Differentially expressed genes in 4th_Ovary versus 3rd_Ovary, 4th_Ovary versus 2rd_Ovary, and 4th_Ovary versus 1st_Ovary (FDR ≤ 0.001 AND |log2 ratio| ≥ 1). (XLSX 14929 KB)
Rights and permissions
About this article
Cite this article
Zhao, C., Wang, P. & Qiu, L. RNA-Seq-based transcriptome analysis of reproduction- and growth-related genes in Lateolabrax japonicus ovaries at four different ages. Mol Biol Rep 45, 2213–2225 (2018). https://doi.org/10.1007/s11033-018-4383-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4383-5