Abstract
Background
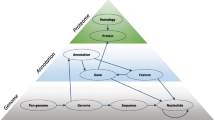
The availability of multiple, essentially complete genome sequences of prokaryotes and eukaryotes spurred both the demand and the opportunity for the construction of an evolutionary classification of genes from these genomes. Such a classification system based on orthologous relationships between genes appears to be a natural framework for comparative genomics and should facilitate both functional annotation of genomes and large-scale evolutionary studies.
Results
We describe here a major update of the previously developed system for delineation of Clusters of Orthologous Groups of proteins (COGs) from the sequenced genomes of prokaryotes and unicellular eukaryotes and the construction of clusters of predicted orthologs for 7 eukaryotic genomes, which we named KOGs after euk aryotic o rthologous g roups. The COG collection currently consists of 138,458 proteins, which form 4873 COGs and comprise 75% of the 185,505 (predicted) proteins encoded in 66 genomes of unicellular organisms. The euk aryotic o rthologous g roups (KOGs) include proteins from 7 eukaryotic genomes: three animals (the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster and Homo sapiens), one plant, Arabidopsis thaliana, two fungi (Saccharomyces cerevisiae and Schizosaccharomyces pombe), and the intracellular microsporidian parasite Encephalitozoon cuniculi. The current KOG set consists of 4852 clusters of orthologs, which include 59,838 proteins, or ~54% of the analyzed eukaryotic 110,655 gene products. Compared to the coverage of the prokaryotic genomes with COGs, a considerably smaller fraction of eukaryotic genes could be included into the KOGs; addition of new eukaryotic genomes is expected to result in substantial increase in the coverage of eukaryotic genomes with KOGs. Examination of the phyletic patterns of KOGs reveals a conserved core represented in all analyzed species and consisting of ~20% of the KOG set. This conserved portion of the KOG set is much greater than the ubiquitous portion of the COG set (~1% of the COGs). In part, this difference is probably due to the small number of included eukaryotic genomes, but it could also reflect the relative compactness of eukaryotes as a clade and the greater evolutionary stability of eukaryotic genomes.
Conclusion
The updated collection of orthologous protein sets for prokaryotes and eukaryotes is expected to be a useful platform for functional annotation of newly sequenced genomes, including those of complex eukaryotes, and genome-wide evolutionary studies.
Similar content being viewed by others
Background
The rapid accumulation of genome sequences is a major challenge to researchers attempting to extract the maximum functional and evolutionary information from the new genomes. To avoid informational overflow from the constant influx of new genome sequences, a comprehensive evolutionary classification of the genes from all sequenced genomes is required. Such classifications are based on two fundamental notions from evolutionary biology: orthology and paralogy, which describe the two fundamentally different types of homologous relationships between genes [1–4]. Orthologs are homologous genes derived by vertical descent from a single ancestral gene in the last common ancestor of the compared species. Paralogs, in contrast, are homologous genes, which, at some stage of evolution of the respective gene family, have evolved by duplication of an ancestral gene. The notions of orthology and paralogy are intimately linked because, if a duplication (s) occurred after the speciation event that separated the compared species, orthology becomes a relationship between sets of paralogs (co-orthologs), rather than individual genes. A classic case of the interplay between orthologous and paralogous relationships is seen in the globin family: all animal globins, including myoglobin, are paralogs, but they are all co-orthologs of the plant leghemoglobin(s) [5].
Deciphering orthologous and paralogous relationships among genes is critical for both the functional and the evolutionary aspects of comparative genomics [4, 5]. Orthologs typically occupy the same functional niche in different species, whereas paralogs tend to evolve toward functional diversification. Therefore, robustness of genome annotation depends on accurate identification of orthologs. Similarly, knowing which homologous genes are orthologs and which are paralogs is required for constructing evolutionary scenarios involving, along with vertical inheritance, lineage-specific gene loss and horizontal gene transfer.
In principle, identification of orthologs requires phylogenetic analysis of entire families of homologous proteins, which is expected to isolate orthologous protein sets in distinct clades [6–8]. However, on the scale of complete genomes, such analysis is both extremely labor-intensive and error-prone due to the inherent artifacts of phylogenetic tree construction. Therefore shortcuts have been developed by introducing the notion of a genome-specific best hit (BeT). A BeT is the protein in a target genome, which is most similar to a given protein from the query genome [9, 10]. The underlying premise is that orthologs are more similar to each other than they are to any other protein from the respective genomes. In multiple-genome comparisons, pairs of potential orthologs identified via BeTs can be joined to form clusters of orthologs represented in all or a subset of the analyzed genomes [9, 11]. This approach to the identification of orthologous protein sets meets with two obvious complications. Firstly, many proteins belong to lineage-specific expansions, i.e., have evolved via duplication(s) after the divergence of the compared species [12–14]. In these cases, deciphering (co)orthologous relationships can be a hard task and clusters of orthologs that include such expansions should be treated with particular caution. The second complication is caused by the fact that many proteins exist in multidomain forms encoded by a single gene in some species and as products of two or more stand-alone genes in others. In protein clustering, multidomain proteins may connect distinct clusters of orthologs resulting in artifactual lumping.
The approach to the identification of orthologous protein sets based on clustering of consistent BeTs has been implemented in the collection of Clusters of Orthologous Groups (COGs) of proteins [9, 15]. The COG construction protocol included an automatic procedure for detecting candidate sets of orthologs, manual splitting of multidomain proteins into the component domains, and subsequent manual curation and annotation. The COGs started with 6 prokaryotic genomes and one genome of a unicellular eukaryote, yeast Saccharomyces cerevisiae [9]. Subsequent updates increased the number of prokaryotic genomes in the COGs to 43 [15]. The procedure for COG construction required that each COG included proteins from at least three sufficiently distant species. This conservative approach notwithstanding, ~60 to ~85% of the proteins encoded in prokaryotic genomes were included in the COGs.
The COG system, which includes the COGNITOR program for adding new members to COGs (RLT, unpublished results), has become a widely used tool for computational genomics. The most important applications of the COGs are functional annotation of newly sequenced genomes [16–20] and genome-wide evolutionary analyses [21–25].
Here, we present a major update to the COGs, with over 63 sequenced prokaryotic genomes and three genomes of unicellular prokaryotes now included. Furthermore, the COG system is extended to complex, multicellular eukaryotes by constructing clusters of probable orthologs, which we named KOGs (euk aryotic o rthologous g roups) for 7 sequenced genomes of animals, fungi, microsporidia, and plants.
Results and discussion
Update of the COGs
To add a new species to the COG system, the annotated protein sequences from the respective genome were compared to the proteins in the COG database by using the BLAST program and assigned to pre-existing COGs by using the COGNITOR program (and see Materials and Methods). The genomes of prokaryotes and unicellular eukaryotes that have been sequenced since the latest update of the COGs were added one at a time. At each step, the proteins that remained unassigned after manual validation of the COGNITOR results were subject to the COG construction procedure in order to identify new COGs that could be formed thanks to the addition of the analyzed genome. The resulting COG assignments for 63 prokaryotic genomes and three genomes of unicellular eukaryotes are quantified in Table 1. The addition of new species leads to incremental increase in the COG coverage for each of the included prokaryotic genomes. The highest coverage now achieved is for Buchnera sp. (99%) and the lowest coverage is for Borrelia burgdorferi (43%). Each of these organisms is a special case. Buchnera is a highly degraded ensymbiont, which evolved from a relatively recent common ancestor with E. coli but apparently lost the great majority of genes, retaining – almost exclusively – conserved, essential ones [26], whereas Borrelia has numerous plasmids that mostly encode poorly conserved genes [27]. Probably more telling is the observation that, for most free-living prokaryotes, ~80% of the genes belong to COGs and there is no appreciable dependence between the number of genes in a genome and the COG coverage (Table 1). Given that most genomes encode a substantial fraction (up to 10%) of fast-evolving, non-globular proteins [28] and other poorly conserved proteins (e.g., remnants of prophages) as well, these findings seem to suggest that the COG coverage of most genomes is approaching saturation.
The COGs are accompanied by a phyletic pattern search tool, i.e., a Web-based tool that allows the user to select COGs with a desired pattern of presence-absence of species. Using the phyletic pattern search tool, one can classify the COGs by the representation of the major lineages of unicellular life forms (Fig. 1). This breakdown of the updated COGs emphasizes the important trend noticed previously[9, 15]: only a minuscule fraction (~1%) of the COGs are ubiquitous and even the COGs that are present in all bacteria or in all archaea represent a small minority. Furthermore, many COGs show scattered distribution, which appears to reflect rampant lineage-specific gene loss and horizontal gene transfer, which are typical of prokaryotic evolution [29–31].
Phyletic patterns of COGs. All, represented in all unicellular organisms included in the COG system; All archaea, All bacteria, All eukaryotes, represented in each species from the respective domain of life (and possibly in some species from other domains); All bacteria except the smallest, represented in all bacteria except, possibly, parasites with small genomes (mycoplasma, chlamydia, rickettsia, and spirochetes).
Construction of KOGs for 7 sequenced eukaryotic genomes
Eukaryotic KOGs were constructed from annotated proteins encoded in the genomes of three animals (Homo sapiens [32], the fruit fly Drosophila melanogaster [33], and the nematode Caenorhabditis elegans) [34], the green plant Arabidopsis thaliana (thale cress) [35], two fungi (budding yeast Saccharomyces cerevisiae [36] and fission yeast Schizosaccharomyces pombe [37], and the microsporidian Encephalitozoon cuniculi [38]). The basic procedure for KOG construction was the same as the procedure previously employed for prokaryotic genomes (Refs. [9, 15] and see Materials and Methods). Given the abundance of multidomain architectures among eukaryotic proteins and the fact that apparent orthologs often differ in domain composition [32, 39], the protocol based on the BeT analysis was amended with domain identification using the RPS-BLAST program [40]. Proteins assigned to a KOG by the initial KOG construction procedure were kept in that KOG without splitting them into individual domains if they shared a common core of domains. In addition, proteins, which consisted solely of widespread, "promiscuous" domains (e.g., SH2, SH3, WD40 repeats or TPR repeats) and did not show clear-cut orthologous relationships, were assigned to Fuzzy Orthologous Groups (FOGs). In addition to KOGs and FOGs, we also identified provisional clusters of orthologs represented in two genomes (TWOGs) by detecting bi-directional BeTs between proteins not included in KOGs or FOGs and assigning additional members by examination of the BLAST search outputs. Finally, lineage-specific expansions (LSEs) of paralogs among the proteins from each genome not included in KOGs, FOGs, and TWOGs were detected by using the clustering procedure described previously [14] accompanied by a newly developed procedure for finding tight protein clusters (BK and RLT, unpublished results). The construction of TWOGs and LSEs involved more extensive case by case evaluation than the KOG construction due to the lack of well established procedures to generate these types of clusters; nevertheless, these clusters should be considered preliminary until further validation.
Table 2 shows the assignment of the proteins from each of the analyzed eukaryotic species to KOGs. Unlike the situation with prokaryotic COGs (Table 1), the fraction of proteins assigned to KOGs tends to decrease with increasing genome size of the analyzed eukaryotic species, from the maximum of ~74% for fission yeast Schizosaccharomyces pombe, the second smallest genome (for reasons that remain unclear, the smallest genome, that of the microsporidian Encephalitozoon cuniculi, had only 61% of the proteins included in COGs) to ~49% for the largest, human genome (Table 2).
Compared to prokaryotes, a considerably smaller fraction of eukaryotic genes could be included into KOGs (Tables 1 and 2). Thus, the apparent difference in coverage with highly conserved clusters of orthologs (C/KOGs) between prokaryotes and eukaryotes, particularly complex ones, is probably due to the relatively small number of eukaryotic genomes included in this analysis and is expected to level off with the growth of the eukaryotic genome collection. This view is compatible with the observed dependence of the KOG coverage on the number of genes (Table 1), which suggests that the KOGs are still far from saturation.
Examination of the phyletic patterns of KOGs points to the existence of a conserved eukaryotic gene core as well as substantial diversity (Fig. 2); this clearly resembles the evolutionary pattern seen previously during the analysis of archaeal COGs [41]. The genes represented in each of the 7 analyzed genomes comprise ~20% of the KOG set and approximately the same number of KOGs includes 6 species, with the exception of the microsporidian. The prevalence of the latter pattern is not surprising given that microsporidia are intracellular parasites with minimal metabolic capabilities and a dramatically reduced genome [38]. The next largest group consists of animal-specific COGs, which, again, could be expected because animals are the only lineage of complex eukaryotes that is represented by more than one species in the analyzed set of genomes. However, a notable observation is that ~30% of the KOGs had "odd" phyletic patterns, e.g., are represented in one animal, one plant and one fungal species (Fig. 2).
Phyletic patterns of KOGs. All, include representatives from each of the 7 analyzed species; All-Ec, include representatives from each of 6 species other than Encephalitozoon cuniculi; All animals, include representatives from three animal genomes only; All fungi, include representatives from two fungal genomes only.
To illustrate the typical composition of a KOG, some of the problems that tend to emerge with their construction, and possible biological implications, we briefly discuss here KOG3378, which includes proteins already mentioned above as a typical case of paralogy and orthology, namely, the globins (Fig. 3). Globins are small (typically, between 140 and 150 amino acid residues) and relatively poorly conserved proteins. As a consequence, the initial, automatic procedure for KOG construction produced a candidate KOG consisting of only 3 proteins from 3 species: S. cerevisiae YGR234w, its ortholog from S. pombe SPAC869.02c, and human neuroglobin Hs10864065. The remaining proteins were brought into the KOG manually, as the result of examination of BLAST search outputs, focused on the conservation of the globin-specific sequence motifs. The final KOG is represented in 6 of the 7 analyzed eukaryotic species, with the sole exception of E. cuniculi (Fig. 3). The most remarkable aspect of this KOG is the apparent independent proliferation of genes for globins and globin-like proteins in vertebrates (represented here by humans): 11 paralogs, and nematodes (C. elegans): 24 paralogs (CE23430 and CE23431 are parts of the same gene). Strictly speaking, to demonstrate that these expansions are, indeed, independent, rather than ancestral, complete phylogenetic analysis is required, which is a difficult task given the low sequence conservation in many members of the KOGs. However, the presence of only one globin homolog in D. melanogaster is best compatible with hypothesis of lineage-specific expansion because, regardless of the exact topology of the animal phylogenetic tree [42], the alternative to this hypothesis would involve massive loss of globin-like genes in insects. Furthermore, this hypothesis is also compatible with the topology of the crude similarity dendrogram, which accompanies the KOG and in which the majority of human and nematode members form distinct clusters (Fig. 3). Thus, at this stage, the most likely, conservative interpretation of the evolutionary relationship between vertebrates and nematode globins is that they comprise co-orthologous sets and are legitimately included in the same KOG. Similarly, the two paralogous leghemoglobins from A. thaliana should be considered co-orthologous to the human and C. elegans paralogous sets.
An example of a complex eukaryotic KOG: globins and related hemoproteins. The systematic protein names of the KOG members are listed under each species. To the left of the KOG proper is the similarity dendrogram produced from the BLAST scores between the KOG members. This is a crude clustering, which should not be construed as a phylogenetic tree.
The functions of human globins and globin homologs, primarily in oxygen delivery to different tissues, at different developmental stages have been studied in great detail [43]. In contrast, the dramatic proliferation of globin-like proteins in the nematode C. elegans, while noticed, in part, in previous work [44], is not well understood. To our knowledge, KOG3378 is the most complete current representation of this lineage-specific expansion of globin-like paralogs; the experimental study of these genes is expected to reveal novel aspects of invertebrate physiology.
Another notable observation comes from the analysis of the yeast members of KOG3378. A BLAST search of the non-redundant protein sequence database (NCBI, NIH, Bethesda) and examination of the domain composition of the S. cerevisiae protein YGR234w shows that this protein (named flavohemoglobin) consists of the globin domain fused to a flavodoxin reductase domain and is highly similar to a variety of oxidoreductases from several bacterial species and some lower eukaryotes (e.g., slime molds and other protists), which have the same domain composition ([45] and data not shown). The S. pombe flavohemoglobin belongs to the same protein family but is not the closest relative of the S. cerevisiae flavohemoglobin (data not shown). These observations strongly suggest that the yeast flavohemoglobin genes have been acquired from bacteria via horizontal gene transfer and hence have an evolutionary history that is distinct and independent from those of plant and animal globins. Notably, the second member of this KOG from S. cerevisie YNL234w is not at all a close paralog of the flavohemoglobins. The only identifiable domain in this large protein is the globin domain, which is most similar to vertebrate neuroglobins. These observations illustrate an important general point to be kept in mind when perusing the KOGs: although a given set of proteins may have been legitimately brought together in the same KOG in the context of eukaryotic genome comparison, on some occasions, different KOG members have different evolutionary trajectories.
Prokaryotic and eukaryotic orthologous gene sets: evolutionary connections and functional differences
The two sets of orthologous genes overlap because the three species of unicellular eukaryotes were included in both sets; the proteins from these species obviously form connections between prokaryotic orthologous sets (COGs) and eukaryotic orthologous sets (KOGs). Such connections, suggestive of orthologous relationships, were established between 1253 COGs, each of which included at least one protein from a unicellular eukaryote (not counting COGs that consisted exclusively of eukaryotic proteins), and 2000 eukaryotic KOGs. The greater number of eukaryotic KOGs involved in this relationship is due to the fact that, on many occasions, several proteins from unicellular eukaryotes that are part of the same COG have their distinct orthologs in other eukaryotes and, accordingly, belong to several KOGs. Only relatively small fractions of the prokaryotic COGs (27% of the COGs that include at least one prokaryotic species) and eukaryotic KOGs (34% of the KOGs and TWOGs) comprised sets of putative orthologs represented in both prokaryotes and eukaryotes. This emphasizes the distinction between the repertoires of genes that are conserved in prokaryotes and in eukaryotes and the considerable amount of innovation in both groups of organisms. However, these numbers give the low bound of the shared clusters of orthologs because some of the KOGs are not represented in the relatively small genomes of unicellular eukaryotes, primarily due to gene loss in the latter, but have prokaryotic counterparts.
Functional annotation of the detected orthologous clusters is one of the crucial and most labor-consuming aspects of the C/KOG analysis. Given the well-known inaccuracy of the currently available schemes for automatic annotation (e.g., Refs. 5,18, and references therein), no attempt was made to fully automate the C/KOG annotation; instead, assignments were made on a case by case basis through a combination of published data on C/KOG members and their homologs, protein domain analysis and different types of context analysis, particularly phyletic patterns and, in prokaryotes, conservation of gene strings which comprise putative operons [46–48]. Figure 4 shows the distribution of known and predicted protein functions for the prokaryotic COGs (i.e., the subset of the COGs obtained by subtraction from the COG collection of those COGs that included solely unicellular eukaryotes were) and the eukaryotic KOGs. The difference between prokaryotic and eukaryotic clusters of orthologs is obvious in that the latter are substantially enriched in proteins involved in signal transduction and intracellular trafficking; certain functional categories, such as cytoskeleton formation and chromatin dynamics were unique to eukaryotes. In contrast, metabolic and transport functions were relatively more prominent among the prokaryotic COGs (Fig. 4).
Functional classification of prokaryotic (COGs) and eukaryotic (KOGs) clusters of orthologs. Designations of functional categories: A, RNA processing and modification (not used for prokaryotic COGs), B, chromatin structure and dynamics, C, energy production and conversion, D, cell cycle control and mitosis, E, amino acid metabolism and transport, F, nucleotide metabolism and transport, G, carbohydrate metabolism and transport, H, coenzyme metabolism, I, lipid metabolism, J, translation, K, transcription, L, replication and repair, M, cell wall/membrane/envelope biogenesis, N, Cell motility, O, post-translational modification, protein turnover, chaperone functions, P, Inorganic ion transport and metabolism, Q, secondary metabolites biosynthesis, transport and catabolism, T, signal transduction, U, intracellular trafficking and secretion, Y, nuclear structure (not applicable to prokaryotic COGs), Z, cytoskeleton (not applicable to prokaryotic COGs); R, general functional prediction only (typically, prediction of biochemical activity), S, function unknown. The numbers were obtained after subtracting the COGs that consisted entirely of proteins from unicellular eukaryotes from the COG collection.
Using phyletic patterns to examine gene function and evolution
Phyletic pattern search can be employed for preliminary assessment of specific functional and evolutionary hypotheses. With the increased number of included genomes and enhanced capabilities of the phyletic pattern search tool, this analysis becomes particularly informative. Below we discuss straightforward examples of its use. Figure 5a shows the results of querying the COG database for COGs that are represented in the microsporidian parasite Encephalitozoon cuniculi but not in the two yeast species. Given the dramatic genome reduction seen in the microsporidium [38], it is not unexpected that the query retrieves only a small set of 13 COGs. This phyletic pattern can be explained either by loss of ancestral eukaryotic genes in yeast or by acquisition of genes by E. cuniculi via horizontal gene transfer. At least in some cases, further examination of the phyletic patterns of the retrieved COGs suggests the most likely scenario. Thus, COGs 1078, 1258, 1690, and 2263 are represented in all archaea, but are either missing in bacteria or are present in a minority of species (Fig. 5a). Therefore these COGs most likely are part of the ancestral archaeo-eukaryotic heritage [49] and might have been lost in yeasts; the respective proteins are known or predicted to be involved in translation or RNA modification, which is compatible with this evolutionary scenario. In contrast, COG3202 seems to be a likely case of horizontal gene transfer. Remarkably, the proteins in this COG are ADP/ATP translocases, which seem to be a hallmark of intracellular parasitism (or symbiosis) allowing the respective organisms to tap into the ATP supplies of the host cell [50, 51]. Indeed, this COG is shared by the eukaryotic (E. cuniculi) and bacterial (Chlamydia and Rickettsia) intracellular parasites, the only exception being a diverged member of the COG found in the plant pathogenic bacterium Xylella fastidiosum (Fig. 5a).
Examples of phyletic pattern search. (A) COGs represented in Encephalitozoon cuniculi but missing in the two yeasts (B) COGs represented in Yersinia pestis but not in other Proteobacteria or eukaryotesThe sets of species included in COGs are color-coded as follows (from left to right): yellow, archaea; purple, eukaryotes; green, miscellaneous bacteria, including hyperthermophiles, cyanobacteria, Fusobacterium, and Deinococcus; dark yellow, actinobacteria; torqoise, low-GC Gram-positive bacteria (except for mycoplasmas); light blue, Gamma-proteobacteria; dark-blue, Beta- and Epsilon-proteobacteria; dark gray, Alpha-proteobacteria; green, chlamydia and spirochetes; dark green, mycoplasmas. The functional categories, designated as in Fig. 4, are also color-coded.
The second case in point that we consider here is a search for COGs, which are represented in the causative agent of plague, Yersinia pestis [52], but not in other Proteobacteria (the taxon to which Y. pestis belongs) or eukaryotes; this query retrieves 7 COGs (Fig. 5b). These genes probably have been acquired by Y. pestis via horizontal gene transfer. On a more practical note, some of these genes could be potential targets for highly selective anti-bacterial agents. It is noticeable that three of these genes are predicted to be involved in cell wall metabolism (COGs 2152, 2401, and 3867), whereas the functions of others remain uncharacterized.
Conclusions
The collection of COGs from prokaryotes and unicellular eukaryotes was substantially amended to include 66 species and eukaryotic orthologous groups (KOGs) for 7 species were constructed. The prokaryotic COG system already covers most of the globular proteins encoded in bacterial and archaeal genomes. Eukaryotic KOGs include a lower fraction of the encoded proteins but this difference is expected to level off with the growth of the eukaryotic genome collection. The eukaryotic KOG analysis revealed a substantial conserved core of eukaryotic genes as well as major lineage-specific variations. Lineage-specific expansion of paralogous families within the KOGs and expansion of families that do not have orthologs in other compared genomes make major contributions to the eukaryotic gene repertoire. Only a minority of eukaryotic KOGs have readily detectable prokaryotic counterparts and the same holds for prokaryotic COGs, emphasizing the extent of innovation in both the eukaryotic and prokaryotic divisions of life. The wide scatter of the phyletic patterns among the KOGs testifies to the importance of lineage-specific gene loss in the evolution of eukaryotic genomes.
The current collection of eukaryotic KOGs includes 7 genomes whose sequences had been available as of July 1, 2002. Manual correction and annotation of KOGs is a labor-intensive process, which precluded immediate inclusion of the genomes of the mouse [53], fugu fish [54], mosquito [55], the urochordate Ciona instestinalis [56], and the malarial parasite Plasmodium falciparum [57], which have become available since that date. However, once the basic system is established, it is expected that inclusion of these and other newly sequenced genomes in the KOG system proceeds at a greater pace.
The C/KOG system can be employed for functional annotation of genes from new genomes by using the COGNITOR program and for research into genome evolution. The utility of the system for both of these purposes should increase progressively with the inclusion of new genomes, particularly those of early-branching eukaryotes.
Methods
Protein sets for new genomes
The protein sets for all newly included bacterial and archaeal genomes, the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, the microsporidian Encaephalitozoon cuniculi, the thale cress Arabidopsis thaliana, and the fruit fly Drosophila melanogaster were extracted from the Genome division of the (NCBI, NIH, Bethesda). The protein sequences for the nematode Caenorhabditis elegans were from the WormPep67 database, the sequences for Homo sapiens were from the NCBI build 30.
Addition of new genomes to the COGs
The new genomes were added to the COGs by using the COGNITOR program, with the results validated manually, essentially as described previously [9, 15]. After the completion of the validation process, the remaining proteins were subject to the COG construction procedure, in order to detect new COGs that could not be formed without the added genomes; the validation and annotation steps were repeated with the newly detected COGs.
Sequence analysis, construction and annotation of KOGs
The construction of KOGs followed the previously outlined strategy based on sets of consistent BeTs [9, 15], but included additional steps that reflected specific features of eukaryotic proteins. Briefly, the procedure was as follows. 1. Detection and masking of widespread, typically repetitive domains, which was performed by using the RPS-BLAST program and the PSSMs for the respective domains from the CDD collection [40]. These domains, namely, PPR (pfam01535), WD40 (pfam00400), IG (pfam00047), IGc1, Igv, IG_like, RRM (pfam00076), ANK (pfam00023), myosin tail (pfam01576), Fn3 (pfam00041), CA, (IG), ANK, kelch (pfam01344), OAD_kelch, SH3 (pfam00018), intermediate filaments (pfam00038), C2H2 finger (pfam00096), PDZ (pfam00595), POZ (pfam00651), PH (pfam00169), ZnF-C4 (pfam00105), spectrin (pfam00435), Sushi (pfam00084), TPR (pfam00017), BTB, LRR_CC, LY, ARM, SH2, and CH, were detected and masked prior to applying the COG construction procedure. Masking these domains was required to ensure the robust classification of the eukaryotic orthologous clusters with the KOG detection procedure because hits between these common, "promiscuous" domains resulted in spurious lumping of numerous non-orthologous proteins. 2. All-against-all comparison of protein sequences from the analyzed genomes by using the gapped BLAST program [58], with filtering for low sequence complexity regions performed using the SEG program [59]. 3. Detection of triangles of mutually consistent, genome-specific best hits (BeTs). 4. Merging triangles with a common side to form crude, preliminary KOGs. 5. Case by case analysis of each candidate KOG. This analysis serves to eliminate the false-positives that are incorporated in the KOGs during the automatic steps and included, primarily, examination of the domain composition of KOG members, which was determined using the RPS-BLAST program and the CDD collection of position-specific scoring matrices (PSSMs) for individual domains [40]. Generally, proteins were kept in the same KOG when they shared a conserved core domain architecture. However, in cases when KOGs were artificially bridged by multidomain proteins, the latter were split into individual domains (or arrays of domains) and steps (1)-(4) were repeated with these sequences; this results in the assignment of individual domains to KOGs in accordance with their distinct evolutionary affinities. 6. Assignment of proteins containing promiscuous domains. In cases when a sequence assigned to a KOG contained one or more masked promiscuous domains, these domains were restored and became part of the respective KOG. Proteins containing promiscuous domains but not assigned to any KOG were classified in Fuzzy Orthologous Groups (FOGs) named after the respective domains. 7. Examination of large KOGs, which included multiple members from all or several of the compared genomes by using phylogenetic trees, cluster analysis with the BLASTCLUST program ftp://ftp.ncbi.nih.gov/blast/, comparison of domain architectures, and visual inspection of alignments; as a result, some of these protein sets were split into two or more smaller ones that were included in the final set of KOGs.
The KOGs were annotated on the basis of the annotations available through GenBank and other public databases, which were critically assessed against the primary literature. For proteins that are currently annotated as "hypothetical" or "unknown", iterative sequence similarity searches with the PSI-BLAST program [58], the results of the RPS-BLAST searches, additional domain architecture analysis performed by using the SMART system [60], and comparison to the COG database by using the COGNITOR program (RLT, unpublished results) were employed to identify distant homologs with experimentally characterized functions and/or structures. The known and predicted functions of KOGs were classified into 23 categories (see legend to Fig. 4); these were modified from the functional classification previously employed for prokaryotic COGs [15] by including several specific eukaryotic categories.
Availability of the results
The updated version of the COGs for unicellular organisms and the eukaryotic KOGs are accessible at http://www.ncbi.nlm.nih.gov/COG/ and via ftp at ftp://ftp.ncbi.nih.gov/pub/COG/.
References
Fitch WM: Distinguishing homologous from analogous proteins. Systematic Zoology 1970, 19: 99–106.
Fitch WM: Homology a personal view on some of the problems. Trends Genet 2000, 16: 227–231. 10.1016/S0168-9525(00)02005-9
Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L: Gene families: the taxonomy of protein paralogs and chimeras. Science 1997, 278: 609–614. 10.1126/science.278.5338.609
Sonnhammer EL, Koonin EV: Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet 2002, 18: 619–620. 10.1016/S0168-9525(02)02793-2
Wilson CA, Kreychman J, Gerstein M: Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol 2000, 297: 233–249. 10.1006/jmbi.2000.3550
Sicheritz-Ponten T, Andersson SG: A phylogenomic approach to microbial evolution. Nucleic Acids Res 2001, 29: 545–552. 10.1093/nar/29.2.545
Zmasek CM, Eddy SR: RIO: Analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics 2002, 3: 14. 10.1186/1471-2105-3-14
Storm CE, Sonnhammer EL: Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics 2002, 18: 92–99. 10.1093/bioinformatics/18.1.92
Tatusov RL, Koonin EV, Lipman DJ: A genomic perspective on protein families. Science 1997, 278: 631–637. 10.1126/science.278.5338.631
Huynen MA, Bork P: Measuring genome evolution. Proc Natl Acad Sci U S A 1998, 95: 5849–5856. 10.1073/pnas.95.11.5849
Montague MG, Hutchison CA 3rd: Gene content phylogeny of herpesviruses. Proc Natl Acad Sci U S A 2000, 97: 5334–5339. 10.1073/pnas.97.10.5334
Jordan IK, Makarova KS, Spouge JL, Wolf YI, Koonin EV: Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res 2001, 11: 555–565. 10.1101/gr.GR-1660R
Remm M, Storm CE, Sonnhammer EL: Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 2001, 314: 1041–1052. 10.1006/jmbi.2000.5197
Lespinet O, Wolf YI, Koonin EV, Aravind L: The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res 2002, 12: 1048–1059. 10.1101/gr.174302
Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV: The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 2001, 29: 22–28. 10.1093/nar/29.1.22
Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV, Belova GI, Aravind L, Natale DA, Rogozin IB, Tatusov RL, Wolf YI, Stetter KO, Malykh AG, Koonin EV, Kozyavkin SA: The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci U S A 2002, 99: 4644–4649. 10.1073/pnas.032671499
Natale DA, Shankavaram UT, Galperin MY, Wolf YI, Aravind L, Koonin EV: Genome annotation using clusters of orthologous groups of proteins (COGs) – towards understanding the first genome of a Crenarchaeon. Genome Biology 2001, 5: RESEARCH0009.
Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J, Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, Smith DR: Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol 2001, 183: 4823–4838. 10.1128/JB.183.16.4823-4838.2001
McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK: Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413: 852–856. 10.1038/35101614
Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ: Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 2001, 65: 44–79. 10.1128/MMBR.65.1.44-79.2001
Jordan IK, Kondrashov FA, Rogozin IB, Tatusov RL, Wolf YI, Koonin EV: Constant relative rate of protein evolution and detection of functional diversification among bacterial, archaeal and eukaryotic proteins. Genome Biol 2001, 2: RESEARCH0053. 10.1186/gb-2001-2-12-research0053
Yanai I, Derti A, DeLisi C: Genes linked by fusion events are generally of the same functional category: A systematic analysis of 30 microbial genomes. Proc Natl Acad Sci U S A 2001, 98: 7940–7945. 10.1073/pnas.141236298
Lecompte O, Ripp R, Puzos-Barbe V, Duprat S, Heilig R, Dietrich J, Thierry JC, Poch O: Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res 2001, 11: 981–993. 10.1101/gr.GR1653R
Koonin EV, Makarova KS, Aravind L: Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol 2001, 55: 709–742. 10.1146/annurev.micro.55.1.709
Jordan IK, Rogozin IB, Wolf YI, Koonin EV: Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res 2002, 12: 962–968. 10.1101/gr.87702. Article published online before print in May 2002
Wernegreen JJ: Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet 2002, 3: 850–861. 10.1038/nrg931
Casjens S: Borrelia genomes in the year 2000. J Mol Microbiol Biotechnol 2000, 2: 401–410.
Koonin EV, Mushegian AR, Galperin MY, Walker DR: Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol 1997, 25: 619–637. 10.1046/j.1365-2958.1997.4821861.x
Snel B, Bork P, Huynen MA: Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res 2002, 12: 17–25. 10.1101/gr.176501
Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV: Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol 2001, 1: 8. 10.1186/1471-2148-1-8
Mirkin BG, Fenner TI, Galperin MY, Koonin EV: Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 2003, 3: 2. 10.1186/1471-2148-3-2
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al.: Initial sequencing and analysis of the human genome. Nature 2001, 409: 860–921. 10.1038/35057062
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al.: The genome sequence of Drosophila melanogaster. Science 2000, 287: 2185–2195. 10.1126/science.287.5461.2185
Consortium TCeS: Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 1998, 282: 2012–2018. 10.1126/science.282.5396.2012
Initiative: TAG Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408: 796–815. 10.1038/35048692
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG: Life with 6000 genes. Science 1996, 274: 563–567. 10.1126/science.274.5287.546
Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, et al.: The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415: 871–880. 10.1038/nature724
Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP: Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414: 450–453. 10.1038/35106579
Koonin EV, Aravind L, Kondrashov AS: The impact of comparative genomics on our understanding of evolution. Cell 2000, 101: 573–576.
Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, Liebert CA, Liu C, Madej T, Marchler GH, Mazumder R, Nikolskaya AN, Panchenko AR, Rao BS, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Vasudevan S, Wang Y, Yamashita RA, Yin JJ, Bryant SH: CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 2003, 31: 383–387. 10.1093/nar/gkg087
Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV: Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res 1999, 9: 608–628.
Hedges SB: The origin and evolution of model organisms. Nat Rev Genet 2002, 3: 838–849. 10.1038/nrg929
Pesce A, Bolognesi M, Bocedi A, Ascenzi P, Dewilde S, Moens L, Hankeln T, Burmester T: Neuroglobin and cytoglobin. Fresh blood for the vertebrate globin family. EMBO Rep 2002, 3: 1146–1151. 10.1093/embo-reports/kvf248
Neuwald AF, Liu JS, Lipman DJ, Lawrence CE: Extracting protein alignment models from the sequence database. Nucleic Acids Res 1997, 25: 1665–1677. 10.1093/nar/25.9.1665
Gardner PR, Gardner AM, Martin LA, Dou Y, Li T, Olson JS, Zhu H, Riggs AF: Nitric-oxide dioxygenase activity and function of flavohemoglobins. sensitivity to nitric oxide and carbon monoxide inhibition. J Biol Chem 2000, 275: 31581–31587. 10.1074/jbc.M004141200
Huynen MJ, Snel B: Gene and context: integrative approaches to genome analysis. Adv Prot Chem 2000, 54: 345–379.
Huynen M, Snel B, Lathe W, Bork P: Exploitation of gene context. Curr Opin Struct Biol 2000, 10: 366–370. 10.1016/S0959-440X(00)00098-1
Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV: Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res 2001, 11: 356–372. 10.1101/gr.GR-1619R
Olsen GJ, Woese CR: Archaeal genomics: an overview. Cell 1997, 89: 991–994.
Winkler HH, Neuhaus HE: Non-mitochondrial ATP transport. Trends Biochem Sci 1999, 24: 64–68. 10.1016/S0968-0004(98)01334-6
Wolf YI, Aravind L, Koonin EV: Rickettsiae and Chlamydiae: evidence of horizontal gene transfer and gene exchange. Trends Genet 1999, 15: 173–175. 10.1016/S0168-9525(99)01704-7
Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG: Genome sequence of Yersinia pestis, the causative agent of plague. Nature 2001, 413: 523–527. 10.1038/35097083
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, et al.: Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420: 520–562. 10.1038/nature01262
Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S: Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 2002, 297: 1301–1310. 10.1126/science.1072104
Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chatuverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, et al.: The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298: 129–149. 10.1126/science.1076181
Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS: The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 2002, 298: 2157–2167. 10.1126/science.1080049
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B: Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419: 498–511. 10.1038/nature01097
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25: 3389–3402. 10.1093/nar/25.17.3389
Wootton JC, Federhen S: Analysis of compositionally biased regions in sequence databases. Methods Enzymol 1996, 266: 554–571.
Schultz J, Milpetz F, Bork P, Ponting CP: SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 1998, 95: 5857–5864. 10.1073/pnas.95.11.5857
Acknowledgements
We thank L. Aravind, David Lipman, Kira Makarova and Wei Yang for useful discussions, and Igor Garkavtsev for his contributions at the initial stages of the KOG project.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Tatusov, R.L., Fedorova, N.D., Jackson, J.D. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41 (2003). https://doi.org/10.1186/1471-2105-4-41
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2105-4-41