Abstract
Leaf thickness is an important trait in rice (Oryza sativa L.). It affects both photosynthesis and sink-resource efficiency. However, compared to leaf length and length width, reports seldom focused on leaf thickness due to the complicated measurement and minor difference. To identify the quantitative trait loci (QTL) and explore the genetic mechanism regulating the natural variation of leaf thickness, we crossed a high leaf thickness variety Aixiuzhan (AXZ) to a thin leaf thickness variety Yangdao No.6 (YD 6) and evaluated 585 F2 individuals. We further use bulked sergeant analysis with whole-genome resequencing (BSA-seq) to identify five genomic regions, including chromosomes 1, 6, 9, 10, and 12. These regions represented significant allele frequency differentiation between thick and thin leaf thickness among the mixed pool offspring. Moreover, we conducted a linkage mapping using 276 individuals derived from the F2 population. We fine-mapped and confirmed that chromosome 9 contributed the primary explanation of phenotypic variance. We fine-mapped the candidate regions and confirmed that the chromosome 9 region contributed to flag leaf thickness in rice. We observed the virtual cellular slices and found that the bundle sheath cells in YD 6 flag leaf veins are fewer than AXZ. We analyzed the potential regions on chromosome 9 and narrowed the QTL candidate intervals in the 928-kb region. Candidate genes of this major QTL were listed as potentially controlled leaf thickness. These results provide promising evidence that cloning leaf thickness is associated with yield production in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is the most consumable and staple food crop globally, feeding more than 50% of the world’s population (Godfray et al. 2010). After entering the first 20 years of the twenty-first century, global warming and the COVID-19 pandemic crisis have become severe contradictions towards population boosts and food supplies (Capell et al. 2020). It is widely recognized that the plant-type improvement is the skeleton for enhancing the yield potential of rice (Khush and Gupta 2013; Qian et al. 2016). In crops, the leaf is the main organ to perform photosynthetic processes providing significant carbon resources. In general, leaf rolling, as well as leaf size, is a critical component in plant architecture morphogenesis that significantly affects crop yield and photosynthetic efficiency (Walter et al. 2009). To investigate the molecular mechanisms that determine leaf rolling and size in rice, numerous researches on quantitative trait loci (QTLs) that contribute to these traits were identified in the past two decades. Several universal complicated regulation networks were revealed, including plant hormone signaling pathways, miRNAs, and transcription factors (Xu et al. 2018).
Leaf morphology consists of leaf length, leaf width, and leaf thickness. The top three rice leaves are the primary source of photosynthetic for grains, and over 50% of the carbohydrate accumulated in rice grains is produced by the flag leaves (Jia et al. 2016). Hence, selecting desirable morphological traits of functional leaves is often a major task for breeders in rice breeding programs. Previous studies reported Narrow Leaf 1(Nal1), NARROW AND ROLLED LEAF 1 (Nrl 1), Nal 2, Nal 3, Nal 8, and Nal 9 focusing on leaf width. In addition, SEMI ROLLED LEAF 1 (SRL 1), CURLY FLAG LEAF 1 (CFL 1), ADAXIALIZED LEAF 1 (ADL1), and LEAF INCLINATION 2 (LC2) conferred to leave morphogenesis (Chen et al. 2019; Cho et al. 2013; Hibara et al. 2009; Hu et al. 2010; Li et al. 2013; Qi et al. 2008; Wu et al. 2011; Xiang et al. 2012; Zhao et al. 2010). Besides, the QTLs associated with leaf morphology and rolling also deepen our understanding of this meaningful trait characterization. Li et al. detected two major QTLs on chromosome 4 and chromosome 8 for flag leaf length using double haploid (DH) population (Li et al. 2000). Another group also used the DH population to study the genotype-environment interaction and mapped seven QTLs on five different chromosomes from chromosomes 1, 2, 3, 4, 6, 9, and 10 associated with the flag leaf length (Yan et al. 1999). Three leaf length QTLs on chromosomes 1, 2, and 4 were identified using IR64 derived introgression lines (Farooq et al. 2010). Jia et al. mapped a total of 19 QTLs for the leaf traits on five chromosomes utilizing recombinant inbred lines (RILs) of 90 lines (Jia et al. 2016). These results showed that although the vital role of leaf traits in rice had attracted significant attention, the characterization and identification of candidate genes are rarely reported.
Leaf thickness is one of the major leaf morphological traits and was considered an essential index in high-yielding cultivation and ideal plant-type breeding (Zhu et al. 2016). It has been investigated that a thick leaf is positively associated with high chlorophyll, leaf nitrogen use efficiency, and photosynthetic enzymes concentration (Murchie et al. 2002; Jinwen et al. 2015). However, due to the difficulty of measurement, none of the leaf thickness QTLs had been identified. Our research group paid attention to the leaf thickness measurement from 2005. An instrument using a non-destructive way to measure leaf thickness was subsequently developed for this important trait in rice (Chen et al. 2007). Furthermore, the thickness of the top three leaves from 208 cultivars was determined, and leaf thickness had a significant positive correlation with leaf length and leaf width. These data suggested that thicker leaf was beneficial to increasing the single leaf area (Liu et al. 2014). The ideal rice architecture theories indicate that thick, short, and erect flag leaf would be the typical characters of leaf morphology for high-yielding in rice (Chen et al. 2005). Similar views were proposed in other theories of rice ideal type model breeding as well (Wang et al. 2019; Dastan et al. 2020). Moreover, some indices such as specific leaf weight (SLW) and specific leaf area (SLA) were utilized to evaluate leaf thickness previously, and several QTLs of SLW- and SLA-associated were discovered (Laza et al. 2006; Zhao et al. 2008; Kanbe et al. 2015). Although these indices are capable of converting from several trait values such as leaf area and leaf dry weight into statistical data to reflect indirect plant leaf phenotypes, it is also possible to bring more significant errors among these measurements. These reasons hindered us from fine-mapping or cloning a major QTL associated with leaf thickness in rice.
The combination of bulked segregant analysis with whole-genome resequencing (BSA-seq) had been developed and successfully applied in yeast for the past decade (Ehrenreich et al. 2010; Wenger et al. 2010). Since then, it has become a popular QTL mapping tool in crop application (Takagi et al. 2013; Yang et al. 2013; Lu et al. 2014; Illa-Berenguer et al. 2015). This method is considered an elegant method to rapidly and effectively identify molecular markers tightly linked to the causal gene from a couple of DNA pools composed of two sets of individuals with distinct or opposite extreme phenotypes (Sahu et al. 2020). With the rapid development of next-generation sequencing (NGS) technologies, it is facilitated to identify genomic regions for target traits using the combination of BSA and whole-genome resequencing of DNA pools by high-throughput genotyping. Based on this concept, several methods have been developed for rapid QTLs identification such as SHOREmap, Next-generation mapping (NGM), MutMap, SNP-ratio mapping (SRM), BSA-seq, and SLAF-seq. (Schneeberger et al. 2009; Austin et al. 2011; Abe et al. 2012a; Lindner et al. 2012; Sun et al. 2013; Takagi et al. 2013). Recently, QTLs associated with partial resistance to rice blast disease and seedling vigor were successfully identified using BSA-seq and verified that QTLs could be successfully detected by BSA-seq even in an F2 generation (Abe et al. 2012b; Takagi et al. 2013).
To uncover the leaf thickness determination component and solve the difficulty of measurements, our research group previously reported a method to measurement leaf thickness in an accurate way (Liu et al. 2014, 2019). Subsequently, we use this method to investigate the genetic basis of flag leaf thickness in rice. During our breeding selection studies, we found that the Indica rice variety Aixiuzhan (AXZ) contained a thicker flag leaf, while another famous Indica cultivar Yangdao No.6 (YD 6) showed thinner flag leaf. These two rice cultivars displayed significant differences in the thickness of flag leaf. We further performed hybridization and further fine-mapping for QTL analysis in this study. BSA-seq detected five candidate chromosome regions for flag leaf thickness. The region from 19.10 to 20.03 Mb on chromosome 9 is probably the major QTL for flag leaf thickness, designated as QTL FLAG LEAF THICKNESS 9 (qFLT9). Then, qFLT9 was further validated, and its candidate region was narrowed down to an interval of 0.928 Mb by QTL fine-mapping. The genetic effect of qFLT9 was also estimated by the near isogenic line (NIL). Potential candidate genes were listed and analyzed. These results provide us novel insight into leaf thickness in rice and provide a valuable resource for rice molecular breeding application.
Results
Analysis of leaf morphology in the F 2 mapping population and their parental cultivars
To study the genetic basis of leaf thickness of flag leaf in rice, we found significant differences in the flag leaf thickness between the parental varieties AXZ and YD 6 (Figs. 1A-C). In comparison with YD 6, AXZ showed higher values of FLT up to 313.34 ± 13.90 µm, while YD 6 exhibited less FLT, and reached 258.69 ± 16.08 µm. We subsequently performed the hybridization and generated the F2 population to observe the distribution of FLT among the F2 population. The variation of FLT in the F2 population is extensive, ranging from 218.44 to 360.68 µm and the medium FLT was 282.92 µm. We observed continuous normal distribution of FLT in 585 F2 progenies, indicating that FLT is a complicated quantitative trait controlled by multiple genes. In summary, we found two rice varieties that have different flag leaf thicknesses, indicating that potential QTL existed conferring to flag leaf thickness.
Morphology of YD 6 and AXZ. A Plant architecture of the parental rice varieties YD 6 and AXZ. Scale bar, 5 cm. B Flag leaves of YD 6 and AXZ. Scale bar, 3 cm. C Average leaf thickness between YD 6 and AXZ. Data are given as the mean ± SD. *P < 0.05; **P < 0.01 compared with the corresponding YD 6 using Student’s t-test
Correlation analysis and BSA-seq
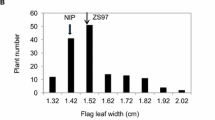
To further identify the major QTL associated with FLT, we created two prototypical pools for BSA-seq. Based on the F2 population frequency distribution, we chose the 218–242-μm flag leaf thickness as L-Bulk, and 327–362-μm flag leaf thickness as H-Bulk (Fig. 2). Thirty individuals with extremely high values of FLT were chosen to an H-bulk. Another 30 individuals with relatively low values were also selected as L-Bulk members as the figure suggested. Genomic DNA of the two bulks was mixed equally to generate bulked DNA and then prepared for DNA sequencing. After the removal of low-quality read and adapter contaminants, a total of 62.56 Gb of sequencing reads were generated from the four DNA pools (Table 1). After aligning all the clean reads to the rice reference genome, a sum of 755,104 variants and SNPs were retained in each pool. These SNPs featured a median read depth of 41–43 in the parents, 36.92 in L-Bulk, and 44.58 in H-Bulk (Table 1).
We further used the program QTLseqr to identify all the differences in allele frequencies between pools and determine 99% credible intervals for QTL mapping. GATK software was applied to analyze the SNPs obtained from comparing the parents and the two F2 pools (Takagi et al. 2013). BSA-seq analysis revealed five genomic regions located in the internal of 32.81–36.97 Mb on chromosomes 1, 26.39–27.57 Mb on chromosomes 6, 19.53–20.76 Mb on chromosomes 9, 11.98–14.11 Mb chromosomes 10, and 0–2.08 Mb on chromosomes 12, which were probably QTLs for flag leaf thickness designated as qFLT1, qFLT6, qFLT9, qFLT10, and qFLT12, respectively (Fig. 3; Table 2). We also displayed BSA-seq results for all 12 chromosomes (Fig. S1). The maximum smoothed allele frequency difference between H-Bulk and L-Bulk was similar in all cases; the QTLs in chromosomes 10 and 12 displayed larger credible intervals, which showed near 2-Mb ranges, while chromosome 6 and chromosome 9 showed narrowed credible intervals just around 1 Mb. These results strongly provided evidence that potential qFLTs exist in these chromosome regions.
Delta SNP index distribution in 12 chromosomes in rice. All different colors represent the SNP-index values after filtering. The black lines indicated the corresponding window SNP-index average values; blue lines indicate 99% credible intervals, and purple lines indicate 95% credible intervals. The windows size is 1 Mb which slide distance is 10 kb. The red circle indicated the candidate intervals on chromosomes. Five potential qFLT positions are also listed in the figure
Validation of BSA-seq results and linkage mapping with F 2 families
To verify the BSA-seq results among these candidate qFLT regions and narrow the potential qFLT credible intervals, we further selected 276 F2 individuals and used Map-QTL to calculate the linkage of disequilibrium (LOD) of these candidate regions (Table S3). However, due to the similarity of AXZ and YD 6, we could not find any applicable InDels (insertions and deletions) and SSRs (simple sequence repeats) in chromosome 1. Therefore, 20 newly designed primers, including InDels and SSRs, were performed to fine-map the candidate positions in chromosomes 6, 9, 10, and 12(Table 3; Table S2). These data showed that qFLT9 has strong LOD values ranging from 6.22 to 11.29. Compared to chromosomes 6, 10, and 12, the LOD values strongly indicated that chromosome 9 is a highly potential candidate QTL that determines flag leaf thickness in rice.
Identification of agronomic traits of parental varieties YD 6, AXZ, and two NILs
Previous studies on gene dominance and recessiveness analysis showed that the loci from YD 6 with thinner leaf thickness are dominant loci in comparison to loci from AXZ. To further explore the association of flag leaf thickness between YD 6 and AXZ, two NILs of the BC2F3 population were constructed. Based on the BSA-seq and F2 fine-QTL mapping results, we successfully generated one population named Thin-NIL (YD 6 allele replaced AXZ allele in AXZ background) and corresponding control as Thick-NIL (AXZ allele in AXZ background). Several agronomic traits, including flag leaf thickness, yield-related traits, and photosynthetic traits, were measured. The flag leaf thickness showed a strong association with NILs in qFLT9 (Fig. 4A1). We found that compared to critical agronomic traits among YD 6 and other materials, the spikelet length, effective grains per panicle, and thousand seed weight and yield per plant showed a significant difference. In contrast, seed setting percentage and yield per plant showed no apparent difference (Fig. 4A2-A6). Other important photosynthetic-related traits, such as soil and plant analyzer development (SPAD) and net photosynthetic rate (Pn), displayed slight differences (Fig. 4A7 and A8). These results suggested that qFLT9 did not contribute to yield-related and photosynthetic-related processes as much as we expected. Furthermore, we utilized XRM (X-ray microscope) to observe the cellular difference among parental plants and Thick-NIL. The results showed that the YD 6 had narrowed side leaf veins (Fig. 5A; Fig. S2A-C), the side leaf vein diameter is approximately 192.23 μm (Fig. 5B). While the side leaf vein in AXZ is about 246.10 μm, near 28% thicker than the YD 6 (Fig. 5C). We also performed the virtual slices observation on the Thick-NIL sample, which showed that the side leaf vein is 242.91 μm, similar to AXZ (Fig. 5C). The cell layer of sieve-tube cell in AXZ and Thick-NIL was near 60% more than that in YD 6 (Fig. S3A). Statistically, we also observed that significant difference in collenchyma cell number and bundle sheath cell number among YD 6, AXZ, and Thick-NIL (Fig. S3A-C). From these results, we could discover that the sieve-tube cells in YD 6 are less than that in AXZ and Thick-NIL, especially when the vertical virtual slices showed that two layers of sieve-tube cells was missing in YD 6 compared to AXZ and THICK-NIL. Other tissues, such as collenchymas, also displayed cell number difference in all three samples (Fig. 5A-C; Fig. S2; Fig. S3).We suspected that the reduction of collenchyma cell number also contributes to leaf thickness reduction in YD 6. In summary, the reduction of sieve-tube cell layer in YD 6 is the primary cellular reason that caused the difference in flag leaf thickness compared to AXZ.
Statistical analysis of YD 6, AXZ, and 2 NILs showed a strong association with qFLT9 and NILs whole plants and leaves phenotypes. A Comparisons of the flag leaf thickness (A1), spikelet length (A2), effective grains per panicle (A3), seed setting percentage (A4), thousand seed weight (A5), yield per plant (A6), SPAD (A7), and Pn (A8). B The plant phenotypes of Thin-NIL and Thick-NIL. Scale bar, 10 cm. C The flag leaf phenotypes of Thin-NIL and Thick-NIL. Scale bar, 2 cm. Data are given as the mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 compared with the corresponding YD 6 using Student’s t-test
Cellular phenotypes of flag leaf in YD 6, AXZ, and Thick-NIL. A Cross-sectional virtual slices of flag leaf in YD 6. (A1) Scale bar, 100 μm. (A1) The yellow rectangle regions are originally from Fig. S2. The A2 showed the yellow line direction vertical virtual splice of A1. There are three sieve-tube cell layers that existed in the leaf vein (green rectangle). Scale bar, 100 μm. The white arrow indicated the reduced sieve-tube cell layers in YD 6. The yellow line measured the diameter of the side leaf vein (A1). The red rectangle indicates the collenchyma, the green rectangle indicates the sieve-tube cells, and the blue shape line shows the bundle sheath in A1 and A2. B Cross-sectional virtual slices of flag leaf in AXZ. (B1) Scale bar, 100 μm. (B1) The yellow rectangle regions are originally from Fig. S2. The B2 showed the yellow line direction vertical virtual splice of B1. Five sieve-tube cell layers and companion cells that existed in the leaf vein (green rectangle). Scale bar, 100 μm. The yellow line measured the diameter of the side leaf vein (B1). The red rectangle indicates the collenchyma, the green rectangle indicates the sieve-tube cells, and the blue shape line shows the bundle sheath, and the yellow rectangle indicates the vessel cell in B1 and B2. C Cross-sectional virtual slices of flag leaf in Thick-NIL. (C1) Scale bar, 100 μm. (C1) The yellow rectangle regions are originally from Fig. S2. The C2 showed the yellow line direction vertical virtual splice of C1. Five sieve-tube cell layers and companion cells existed in the leaf vein (green rectangle). Scale bar, 100 μm. The yellow line measured the diameter of the side leaf vein (C1). The red rectangle indicates the collenchyma, the green rectangle indicates the sieve-tube cells, and the blue shape line shows the bundle sheath, and the yellow rectangle indicates the vessel cell in C1 and C2
To verify our data of qFLT9 in flag leaf thickness determination, we performed a phenotypical measurement of the BC2F4 population among YD 6, AXZ, and the two NILs (Fig. S4). The results showed that the Thick-NIL and Thin-NIL are well associated with the flag leaf thickness in comparison to YD 6 and AXZ. Meanwhile, we further used two BC3F2 populations to fine-map the qFLT9. Three SNP markers and two Indels were utilized to narrow our candidate interval from 2.03 to 0.928 Mb. Based on 20 critical exchanges of single plants, we fine-mapped the qFLT9 intervals between markers L4-3 to L1-3 (Fig. 6). To further verify the existing evidence of qFLT6 and qFLT10, we used BC3F1 single plants and fixed the other genotypes in other potential qFLTs for verification. According to our BC3F2 population results, the qFLT6 and qFLT10 do not show a significant difference in leaf thickness (Table S2, Fig. S5). The results confirmed that the flag leaf thickness is strongly associated with qFLT9, indicating that there is a major QTL that existed in chromosome 9 between 19.10 and 20.03 Mb.
Fine-mapping of qFLT9. Twenty key exchanges were selected from a total 192 BC3F2 population. Two Indels and three SNP markers were used for fine-mapping. The table summarized all 20 exchanges genotypes and phenotypes of leaf thickness. Two different field populations, 12 and 5, were used for the progeny test (each number of plants is 14 and 21, respectively)
Candidate gene identification of qFLT9
The fine-mapping of qFLT9 had narrowed and confirmed that chromosome 9 from 19.10 to 20.01 Mb is the candidate region associated with qFLT9. We considered that the leaf thickness has not only significantly altered between two parental varieties, the leaf length, and leaf width also exhibited differences partially. We also checked the known and published leaf-related QTLs and single mutated genes, no leaf-related genes were fallen into this region so far (Xu et al. 2018) (Table S4). We further analyzed the candidate regions of qFLT9 and listed all the candidate genes below (Table S5). Furthermore, we also listed several most potential genes and their expression patterns. We further chose the 13 most possible candidate genes and listed their expression patterns in the supplemental data (Fig. S6 and 7). In general, 130 candidate genes were listed in these regions, including 41 genes, are annotated as expressed protein, six F-Box transcription factor genes as gene clusters, and three Beta-Glucosidase in a gene cluster. In summary, the candidate genes that determined qFLT9 fell into these potential regions.
Discussion
Plant leaf morphology had been long investigated that associated with yield production and photosynthetic efficiency. Based on the source-sink theory, leaves are the most crucial energy source provider, and the traits focusing on them hugely influence yield development and nutritional quality (Smith et al. 2018). In the past decades, quite many QTLs and single genes that determine leaf morphology have been investigated (Xu et al. 2018), boosting the theories and application in building upregulation networks of source-sink in crops. Leaf length and width-related genes in rice have been studied broadly, while seldom studies that relied on leaf thickness were performed previously. We investigated the leaf thickness by using a leaf thickness measurement machine since last decade (Liu et al. 2014). In this study, we combined traditional fine-mapping methods and high-throughput BSA-seq data; we also narrowed the first leaf thickness QTL in chromosome 9 in rice, filling the research blanks on the leaf morphological determination.
Our studies revealed that the alteration reason of leaf thickness between AXZ and YD 6 is because of the cell layer difference on bundle sheath on the branch veins of flag leaf (Fig. 5). From the BSA-seq data, we scanned five potential intervals associated with flag leaf thickness traits in rice, including chromosomes 1, 6, 9, 10, and 12. Due to the lack of sufficient Indels and SSRs in chromosome 1, we failed to verify the qFLT1 authenticity by the fine-mapping method. Hence, we further used BC3F2 fine-mapping method to verify the other qFLTs and confirmed that the qFLT9 is the major QTLs in flag leaf thickness, and qFLT6 and qFLT10 do not contribute to flag leaf thickness in rice (Fig. S5). However, our data do not exclude the possibility that qFLT12 is also associated with flag leaf thickness (Table S2). Fine-mapping studies towards qFLT12 would be carried out to verify its QTL genetic effect. More studies, such as overexpression and CRISPR-Cas9 gene editing of the candidate genes, would provide more information on qFLT9. After fine-mapping, we are convinced that qFLT9 is the major QTL for flag leaf thickness in rice; subsequently, after verification and clone of qFLT9, we would investigate the human selection evidence. Our NILs and parent varieties agronomic trait statistical analysis also indicated that qFLT9 did contribute to leaf thickness, spikelet length, effective grain per panicle, and thousand seed weight. However, the most important agronomic trait related to yield, that is, yield per plant, did not show a positive association towards qFLT9, indicating that qFLT9 or leaf thickness did not provide a strong novel trade-off to yield production (Fig. 4). This result also partially explained that there was no any single known leaf-related gene in these regions that had been reported before.
To investigate the potential candidate gene of qFLT9, we listed all the possible genes (Table S5). The gene list had excluded re-transposon proteins as well as hypothetical proteins. From the gene list, we found several interesting phenomena related to the candidate genes. First, we observed that an F-box containing gene cluster (From OsFBX 332 to OsFBX 337) existed in this interval. Interestingly, OsFBX332 and OsFBX336 transcriptional expression levels are very similar. Both of them represented higher expression levels in leaf tissue rather than other tissues (Fig. S6). There are about 687 potential F-box proteins in rice controlling the degradation of cellular proteins in rice (Paterson et al. 1993). Some of them encoded phytohormone important signaling transduction regulators, such as TIR1, COI1, GID2, and MAX2 (Tao et al. 2017; Guo et al. 2021; Liu et al. 2021; Patil et al. 2021). Moreover, a small beta-glucosidase cluster is also presented in the candidate region. Additionally, previous studies on Os4BGlu14 indicated that beta-glucosidase in rice functioned as a negative regulator of seed longevity (Ren et al. 2020). In the expression data, LOC_Os09g33690 (Os9bglu32) displayed significantly high expression in the leaf, marking it as the highly potential candidate gene of qFLT9 (Fig. S7). Surprisingly, a critical rice grain size and number determination gene, OsSPL18, was found in the interval of qFLT9 (Yuan et al. 2019). Besides these genes, several mitochondria and chloroplast-related genes are also represented in the interval. Previous studies also uncovered that several genes located in mitochondria contribute to leaf morphological development (Chen et al. 2019; Huang et al. 2020). Disrupting mitochondria’s normal functions would also cause cell development delays as well as cell metabolomics changes. Two mitochondria carrier proteins (LOC_Os09g32200 and LOC_Os09g33470) showed high expression levels in leaf tissues. Based on former studies on leaf morphology, chloroplast mal-function led to leaf morphology alteration (Table S4). These potential genes are worth considering contributing to flag leaf thickness. We would continuously narrow the interval of qFLT9 and eventually clone the candidate gene for further analysis, combining rice genetic analysis, including overexpression and CRISPR/Cas9 gene-editing, to verify the qFLT9 candidate gene. More cellular and molecular functional studies would be carried out for qFLT9 studies.
Leaf morphology contributes to source-sink energetic balance. However, for an extended period, leaf thickness is neglected due to measurement difficulties. Several papers had been published to use SLW to represent leaf thickness, and several QTLs related to SLW in rice had been discovered in chromosome 6 near PSM388 ~ PSM136 position (Jiang et al. 2010; Li et al. 2010). We hope our methods benefit rice leaf morphology and molecular breeding applications, expand to other crops, such as maize and wheat, would also investigate the leaf thickness, and discover more QTLs associated with leaf thickness.
Conclusion
In conclusion, we used BSA-seq to screen four potential genomic regions associated with flag leaf thickness in rice. Three featured alleles were subsequently excluded from the parents by the F2 and NIL population via linkage mapping; the only potential region in chromosome 9 was narrowed to a 928-kb region containing 130 candidate genes (Table S5). We further discussed the potential candidate genes for qFLT9. qFLT9 functioned in bundle sheath cell number difference (Fig. 5). Despite further genetic evidence needed be performed for solid evidence, the application of regional molecular markers would also benefit molecular breeding in rice. Our mapping resolution was high enough to identify strong candidates for flag leaf thickness in rice conclusively. Nevertheless, these results provide an early step towards revealing the genetic and molecular mechanisms underlying leaf thickness in rice. As far as we know, this is the first report of flag leaf thickness QTL in rice. Our discovery provides novel insight into molecular breeding towards ideal phenotype architecture in rice.
Materials and methods
Plant materials and growth conditions
The mapping population of BC2F3, BC2F4 generation and F2 population were developed from the cross of YD 6 and AXZ (both of them are Indica cultivars, YD 6 × AXZ). The plants were cultivated at the experimental field of Guangzhou (113.377426 E, 23.162136 N), Huizhou (113.592206 E, 23.186376 N), and Huangliu (108.798075 E, 18.504724 N) from 2015 to 2020. The plants were spaced by 16.5 cm within a row, and rows were 19.8 cm distance. Field management followed standard agricultural practices.
Flag leaf thickness and other related trait measurement
Previously, a leaf thickness measurement machine was reported for leaf thickness measurement in rice (Liu et al. 2014, 2019). We measured the flag leaf thickness in the center of the flag leaf 120 days after germination (DAG). Each individual measured three replicates. For other agronomic traits, individuals were harvested and all grains were counted for three biological replicates. More than 200 mature spikelets were weighed with an electronic micrometer balance, with the measurements used to calculate a thousand weights of the seeds. We used a multi-function plant measuring instrument MultispeQ (PhotosynQ, USA) to obtain the SPAD. We used Li-6400XT (LI-COR Biosciences, USA) for photosynthetic traits to obtain the relevant data (Rochaix 2011).
Selection of bulks and sequencing of DNA pools
A total of 585 F2 individuals were cultivated and their flag leaf thickness data was measured to separate into two bulks; each bulk contained 30 individuals. After extracting DNA from samples using CTAB methods (Paterson et al. 1993), all DNAs were properly mixed with equal concentration, forming a final DNA pool representing all samples. Together with the parent samples, the four DNA pools (YD 6, AXZ, H-Bulk, and L-Bulk) were submitted to Hengchuang Gene Co. Ltd. for library preparation and sequencing. PCR-free libraries were created by mechanically shearing DNA samples and ligating adapters and PolyA tails, using reagents equivalent to those in the TruSeq library preparation kit (Illumina, San Diego, California). DNA samples were further qualified by agarose gel electrophoresis analysis, Nanodrop detection for purity, and Qubit accurate measurement. The DNA library was diluted to 1 ng/μl and subsequently detected by Agilent 2100. After qualification, BSA-seq was performed by Illumina × 10 pair-end sequencing.
BSA-seq data analysis
Raw reads were filtered and trimmed from both ends using fastq v 0.20.0 with default parameters (Chen et al. 2018). Trimmed reads were then aligned to the Oryza sativa genome (Kawahara et al. 2013) with bwa v 0.7.12 using the MEM algorithm and default parameters (Li and Durbin 2009). SAMtools were used to sort out and index bam files (Li et al. 2009). The minimum alignment mapping quality was set to 20, and the consensus-caller method was used. The results were preserved as VCF files and subsequently filtered with VCFtools version 0.1.17 to remove InDels and SNPs with more than two alleles (Danecek et al. 2011). GATK and SnpEff were used to detect and verify InDels (McKenna et al. 2010; Cingolani et al. 2012). For the visualization of allele frequencies, allele frequency means for each pool were estimated in 1-Mb sliding windows with a 10-Kb increment. To analyze and visualize the SNP frequencies, QTLseqr software was used (Mansfeld and Grumet 2018).
Fine-mapped cloning of qFLT9
The Thin-NIL was generated by hybriding YD 6 × AXZ. Selected from 192 individuals in BC2F3 populations, the most thin leaf thickness line carried YD 6 DNA fragment on chromosome 9. The Thick-NIL was generated from the same populations in BC2F3 populations, the line carried AXZ DNA fragment on chromosome 9 and its phenotype displayed the most thick leaf thickness. The BC3F3 and BC3F4 populations were generated by YD 6 × AXZ. To fine-map the qFLT9, we designed several new molecular markers from predicted SSRs (simple sequence repeats) and InDels. Three SNP markers were identified and performed in sequencing for qFLT9 fine-mapping. The qFLT9 was finally mapped to a 928-kb region on chromosome 9. A 2 × PCR mix was used to amplify the DNA fragments (Tiangen #KT207). Primer sequences are given in Table S1.
X-ray microscopic observation
YD 6 and AXZ flag leaves were collected and fixed in FAA (50% ethanol, 5% glacial acetic acid, 5% formaldehyde) for 12 h at 4 ℃. After the tissue was dehydrated in a graded ethanol series, the samples were thoroughly desiccated in an automated Critical Point Dryer (Leica EM CPD300). Samples were observed with a Zeiss Xradia 515 Versa X-ray microscope. Dragonfly software was used to generate the visual image slices.
Gene expression analysis
qFLT9 candidate gene list was collected through MSU rice annotation database (http://rice.uga.edu/); the candidate gene expression data was collected and analyzed from rice expression database (http://expression.ic4r.org/index) (Xia et al. 2017).
Data Availability
The datasets supporting the conclusions of this article are included within the article.
Code availability
Not applicable.
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012a) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30(2):174–178. https://doi.org/10.1038/nbt.2095
Abe A, Takagi H, Fujibe T, Aya K, Kojima M, Sakakibara H, Uemura A, Matsuoka M, Terauchi R (2012b) OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor Appl Genet 125(4):647–657. https://doi.org/10.1007/s00122-012-1857-z
Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, McCourt P, Guttman DS (2011) Next-generation mapping of Arabidopsis genes. Plant J 67(4):715–725. https://doi.org/10.1111/j.1365-313X.2011.04619.x
Capell T, Twyman RM, Armario-Najera V, Ma JK, Schillberg S, Christou P (2020) Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci 25(7):635–643. https://doi.org/10.1016/j.tplants.2020.04.009
Chen K, Guo T, Li XM, Yang YB, Dong NQ, Shi CL, Ye WW, Shan JX, Lin HX (2019) NAL8 encodes a prohibitin that contributes to leaf and spikelet development by regulating mitochondria and chloroplasts stability in rice. BMC Plant Biol 19(1):395. https://doi.org/10.1186/s12870-019-2007-4
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chen Y-d, Zhang X, Zhou X-q, Chen G-h (2007) Preliminary studies on thickness of nondestructive rice (Oryza sativa L) leaf blade. Agric Sci China 6(7):802–807. https://doi.org/10.1016/s1671-2927(07)60115-1
Chen Y, Wan B, Zhang X (2005) Plant ideotype at heading for super high-yielding rice in double cropping system in south China. Rice Sci 12(2):92–100
Cho SH, Yoo SC, Zhang H, Pandeya D, Koh HJ, Hwang JY, Kim GT, Paek NC (2013) The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol 198(4):1071–1084. https://doi.org/10.1111/nph.12231
Cingolani P, Platts A, le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (austin) 6(2):80–92. https://doi.org/10.4161/fly.19695
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Genomes Project Analysis G (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Dastan S, Ghareyazie B, Teixeira da Silva JA (2020) Selection of ideotype to increase yield potential of GM and non-GM rice cultivars. Plant Sci 297:110519. https://doi.org/10.1016/j.plantsci.2020.110519
Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L (2010) Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464(7291):1039–1042. https://doi.org/10.1038/nature08923
Farooq M, Tagle AG, Santos RE, Ebron LA, Fujita D, Kobayashi N (2010) Quantitative trait loci mapping for leaf length and leaf width in rice cv. IR64 derived lines. J Integr Plant Biol 52(6):578–584. https://doi.org/10.1111/j.1744-7909.2010.00955.x
Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327(5967):812–818. https://doi.org/10.1126/science.1185383
Guo F, Huang Y, Qi P, Lian G, Hu X, Han N, Wang J, Zhu M, Qian Q, Bian H (2021) Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol 229(5):2676–2692. https://doi.org/10.1111/nph.17061
Hibara K, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y (2009) The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev Biol 334(2):345–354. https://doi.org/10.1016/j.ydbio.2009.07.042
Hu J, Zhu L, Zeng D, Gao Z, Guo L, Fang Y, Zhang G, Dong G, Yan M, Liu J, Qian Q (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol 73(3):283–292. https://doi.org/10.1007/s11103-010-9614-7
Huang S, Xin S, Xie G, Han J, Liu Z, Wang B, Zhang S, Wu Q, Cheng X (2020) Mutagenesis reveals that the rice OsMPT3 gene is an important osmotic regulatory factor. The Crop Journal 8(3):465–479. https://doi.org/10.1016/j.cj.2020.02.001
Illa-Berenguer E, Van Houten J, Huang Z, van der Knaap E (2015) Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor Appl Genet 128(7):1329–1342. https://doi.org/10.1007/s00122-015-2509-x
Jia B, Zhao X, Qin Y, Irfan M, Kim T-H, Wang B, Wang S, Sohn J (2016) Identification of quantitative trait loci for leaf traits in rice. Genetika 48(2):643–652. https://doi.org/10.2298/gensr1602643j
Jiang S, Zhang X, Huang C, Xing Y, Zheng X, Xu Z, Chen W (2010) Comparison of genetic linkage map and QTLs controlling flag leaf traits based on F2 and F2: 6 populations derived from Japonica rice. Chin J Rice Sci 24(4):372–378. https://doi.org/10.3969/j.issn.1001-7216.2010.04.007
Jinwen L, Jingping Y, Dongsheng L, Pinpin F, Tiantai G, Changshui G, Wenyue C (2015) Chlorophyll meter’s estimate of weight-based nitrogen concentration in rice leaf is influenced by leaf thickness. Plant Prod Sci 14(2):177–183. https://doi.org/10.1626/pps.14.177
Kanbe T, Sasaki H, Aoki N, Yamagishi T, Ebitani T, Yano M, Ohsugi R (2015) Identification of QTLs for improvement of plant type in rice (Oryza sativaL.) using Koshihikari / Kasalath chromosome segment substitution lines and backcross progeny F2Population. Plant Prod Sci 11(4):447–456. https://doi.org/10.1626/pps.11.447
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (n y) 6(1):4. https://doi.org/10.1186/1939-8433-6-4
Khush GS, Gupta P (2013) Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breeding:n/a-n/ahttps://doi.org/10.1111/pbr.1991
Laza MR, Kondo M, Ideta O, Barlaan E, Imbe T (2006) Identification of quantitative trait loci for δ13C and productivity in irrigated lowland rice. Crop Sci 46(2):763–773. https://doi.org/10.2135/cropsci2005.05.0084
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li R, Zhao S, Miao T, Xu Z, Chen W (2010) QTL Analysis on flag leaf trait in rice. Crops:26–29. https://doi.org/10.16035/j.issn.1001-7283.2010.03.027
Li S, He P, Wang Y, Li H, Chen Y, Zhou K, Zhu L (2000) Genetic analysis and gene mapping of the leaf traits in rice (Oryza sativa L.). Acta Agronomica Sinica 26(3):261–265
Li W, Wu C, Hu G, Xing L, Qian W, Si H, Sun Z, Wang X, Fu Y, Liu W (2013) Characterization and fine mapping of a novel rice narrow leaf mutant nal9. J Integr Plant Biol 55(11):1016–1025. https://doi.org/10.1111/jipb.12098
Lindner H, Raissig MT, Sailer C, Shimosato-Asano H, Bruggmann R, Grossniklaus U (2012) SNP-ratio mapping (SRM): identifying lethal alleles and mutations in complex genetic backgrounds by next-generation sequencing. Genetics 191(4):1381–1386. https://doi.org/10.1534/genetics.112.141341
Liu B, Seong K, Pang S, Song J, Gao H, Wang C, Zhai J, Zhang Y, Gao S, Li X, Qi T, Song S (2021) Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol 231(4):1525–1545. https://doi.org/10.1111/nph.17477
Liu C-g, Zhou X-q, Chen D-g, Li L-j, Li J-c, Chen Y-d (2014) Natural variation of leaf thickness and its association to yield traits in indica rice. J Integr Agric 13(2):316–325. https://doi.org/10.1016/s2095-3119(13)60498-0
Liu C, Chen D, Zhou X, Guo J, Li J, Li L, Chen Y (2019) Association mapping for flag leaf thickness in an indica rice population from South China. Afr J Agric Res 14(25):1058–1072. https://doi.org/10.5897/ajar2018.13826
Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, Sun J, Zhang Z, Weng Y, Huang S (2014) QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor Appl Genet 127(7):1491–1499. https://doi.org/10.1007/s00122-014-2313-z
Mansfeld BN, Grumet R (2018) QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. Plant Genome 11 (2). https://doi.org/10.3835/plantgenome2018.01.0006
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. https://doi.org/10.1101/gr.107524.110
Murchie EH, Hubbart S, Chen Y, Peng S, Horton P (2002) Acclimation of rice photosynthesis to irradiance under field conditions. Plant Physiol 130(4):1999–2010. https://doi.org/10.1104/pp.011098
Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep 11(2):122–127. https://doi.org/10.1007/bf02670470
Patil SB, Barbier FF, Zhao J, Zafar SA, Uzair M, Sun Y, Fang J, Perez-Garcia MD, Bertheloot J, Sakr S, Fichtner F, Chabikwa TG, Yuan S, Beveridge CA, Li X (2021) Sucrose promotes D53 accumulation and tillering in rice. New Phytol. https://doi.org/10.1111/nph.17834
Qi J, Qian Q, Bu Q, Li S, Chen Q, Sun J, Liang W, Zhou Y, Chu C, Li X, Ren F, Palme K, Zhao B, Chen J, Chen M, Li C (2008) Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol 147(4):1947–1959. https://doi.org/10.1104/pp.108.118778
Qian Q, Guo L, Smith SM, Li J (2016) Breeding high-yield superior quality hybrid super rice by rational design. Natl Sci Rev 3(3):283–294. https://doi.org/10.1093/nsr/nww006
Ren RJ, Wang P, Wang LN, Su JP, Sun LJ, Sun Y, Chen DF, Chen XW (2020) Os4BGlu14, a monolignol beta-Glucosidase, negatively affects seed longevity by influencing primary metabolism in rice. Plant Mol Biol 104(4–5):513–527. https://doi.org/10.1007/s11103-020-01056-1
Rochaix JD (2011) Assembly of the photosynthetic apparatus. Plant Physiol 155(4):1493–1500. https://doi.org/10.1104/pp.110.169839
Sahu PK, Sao R, Mondal S, Vishwakarma G, Gupta SK, Kumar V, Singh S, Sharma D, Das BK (2020) Next generation sequencing based forward genetic approaches for identification and mapping of causal mutations in crop plants: a comprehensive review. Plants (Basel) 9 (10). https://doi.org/10.3390/plants9101355
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6(8):550–551. https://doi.org/10.1038/nmeth0809-550
Smith MR, Rao IM, Merchant A (2018) Source-sink relationships in crop plants and their influence on yield development and nutritional quality. Front Plant Sci 9:1889. https://doi.org/10.3389/fpls.2018.01889
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H, Xu C, Song J, Huang L, Wang C, Shi J, Wang R, Zheng X, Lu C, Wang X, Zheng H (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8(3):e58700. https://doi.org/10.1371/journal.pone.0058700
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74(1):174–183. https://doi.org/10.1111/tpj.12105
Tao T, Zhou CJ, Wang Q, Chen XR, Sun Q, Zhao TY, Ye JC, Wang Y, Zhang ZY, Zhang YL, Guo ZJ, Wang XB, Li DW, Yu JL, Han CG (2017) Rice black streaked dwarf virus P7–2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS One 12(5):e0177518. https://doi.org/10.1371/journal.pone.0177518
Walter A, Silk WK, Schurr U (2009) Environmental effects on spatial and temporal patterns of leaf and root growth. Annu Rev Plant Biol 60:279–304. https://doi.org/10.1146/annurev.arplant.59.032607.092819
Wang D, Fahad S, Saud S, Kamran M, Khan A, Khan MN, Hammad HM, Nasim W (2019) Morphological acclimation to agronomic manipulation in leaf dispersion and orientation to promote “Ideotype” breeding: evidence from 3D visual modeling of “super” rice (Oryza sativa L.). Plant Physiol Biochem 135:499–510. https://doi.org/10.1016/j.plaphy.2018.11.010
Wenger JW, Schwartz K, Sherlock G (2010) Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet 6(5):e1000942. https://doi.org/10.1371/journal.pgen.1000942
Wu R, Li S, He S, Wassmann F, Yu C, Qin G, Schreiber L, Qu LJ, Gu H (2011) CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23(9):3392–3411. https://doi.org/10.1105/tpc.111.088625
Xia L, Zou D, Sang J, Xu X, Yin H, Li M, Wu S, Hu S, Hao L, Zhang Z (2017) Rice expression database (RED): an integrated RNA-Seq-derived gene expression database for rice. J Genet Genomics 44(5):235–241. https://doi.org/10.1016/j.jgg.2017.05.003
Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159(4):1488–1500. https://doi.org/10.1104/pp.112.199968
Xu P, Ali A, Han B, Wu X (2018) Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front Plant Sci 9:1528. https://doi.org/10.3389/fpls.2018.01528
Yan J, Zhu J, He C, Benmoussa M, Wu P (1999) Molecular marker-assisted dissection of genotype enviornment interaction for plant type traits in rice. Crop Sci 39(2):538–544
Yang Z, Huang D, Tang W, Zheng Y, Liang K, Cutler AJ, Wu W (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One 8(7):e68433. https://doi.org/10.1371/journal.pone.0068433
Yuan H, Qin P, Hu L, Zhan S, Wang S, Gao P, Li J, Jin M, Xu Z, Gao Q, Du A, Tu B, Chen W, Ma B, Wang Y, Li S (2019) OsSPL18 controls grain weight and grain number in rice. J Genet Genomics 46(1):41–51. https://doi.org/10.1016/j.jgg.2019.01.003
Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20(8):935–947. https://doi.org/10.1038/cr.2010.109
Zhao X-Q, Xu J-L, Zhao M, Lafitte R, Zhu L-H, Fu B-Y, Gao Y-M, Li Z-K (2008) QTLs affecting morph-physiological traits related to drought tolerance detected in overlapping introgression lines of rice (Oryza sativa L.). Plant Sci 174(6):618–625. https://doi.org/10.1016/j.plantsci.2008.03.009
Zhu G, Peng S, Huang J, Cui K, Nie L, Wang F (2016) Genetic improvements in rice yield and concomitant increases in radiation- and nitrogen-use efficiency in middle reaches of Yangtze River. Sci Rep 6:21049. https://doi.org/10.1038/srep21049
Acknowledgements
The authors are grateful to Prof. Zhang Xu, Rice Research Institute, Guangdong Academy of Agricultural Sciences, for critical reviews of this manuscript. Also, the authors thanked Mr. Jiao Longan, Mr. Cao Chunjie, and Ms. Wang Xiaohong from Carl Zeiss (Shanghai) for technical support of X-ray microscopy observation. We thanked the assistance of explanation of cellular difference in leaf thickness by Mr. Zhang Wenhu, agro-biological gene research center, Guangdong Academy of Agricultural Sciences.
Funding
This study was supported by the Guangdong Key Areas Research and Development Program Project (2020B0202090003), Natural Science Foundation of Guangdong Province, China (2017A030313185, 2021A1515011112), and Guangdong Scientific Program Project (2020B1212060047).
Author information
Authors and Affiliations
Contributions
DC and XZ. performed most of the experiments; KC wrote the manuscript; PC. analyzed the data; JG performed part field experiments; CL designed and supervised this study; YC revised the manuscript. All the authors discussed the results and contributed to the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
All authors are consent to publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Da-gang Chen and Xin-qiao Zhou are contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Dg., Zhou, Xq., Chen, K. et al. Fine-mapping and candidate gene analysis of a major locus controlling leaf thickness in rice (Oryza sativa L.). Mol Breeding 42, 6 (2022). https://doi.org/10.1007/s11032-022-01275-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-022-01275-y