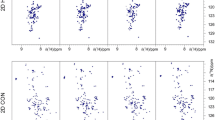
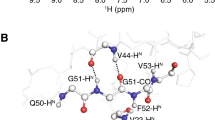
Abstract
NMR assignment of intrinsically disordered proteins (IDPs) by conventional HN-detected methods is hampered by the small dispersion of the amide protons chemical shifts and exchange broadening of amide proton signals. Therefore several alternative assignment strategies have been proposed in the last years. Attempting to seize that dispersion of 13C′ and 15N chemical shifts holds even in IDPs, we recently proposed two 13C-detected experiments to directly correlate the chemical shifts of two consecutive 13C′–15N groups in proteins, i.e. without mediation of other nuclei. Main drawback of these experiments is the interruption of the connection at prolines. Here we present new 13C-detected experiments to correlate consecutive 13C′–15N groups in IDPs, hacacoNcaNCO and hacaCOncaNCO, that overcome this limitation. Moreover, the experiments provide recognition of glycine residues, thereby facilitating the assignment process.



Similar content being viewed by others
References
Aguado-Llera D, Hamidi T, Doménech R, Pantoja-Uceda D, Gironella M, Santoro J, Velázquez-Campoy A, Neira JL, Iovanna JL (2013) Deciphering the binding between Nupr1 and MSL1 and their DNA-repairing activity. PlosOne 8:e78101
Bermel W, Bertini I, Duma L, Emsley L, Felli IC, Pierattelli R, Vasos PR (2005) Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angew Chem Int Ed 44:3089–3092
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2006) 13C detected protonless NMR spectroscopy of proteins in solution. Progr NMR Spectrosc 48:25–45
Bermel W, Felli IC, Kümmerle R, Pierattelli R (2008) 13C direct detection biomolecular NMR. Concepts Magn Reson 32A:183–200
Bermel W, Bertini I, Csizmok V, Felli IC, Pierattelli R, Tompa P (2009a) H-start for exclusively heteronuclear NMR spectroscopy: the case of intrinsically disordered proteins. J Magn Reson 198:275–281
Bermel W, Bertini I, Felli IC, Pierattelli R (2009b) Speeding up 13C direct detection biomolecular NMR experiments. J Am Chem Soc 131:15339–15345
Bermel W, Bertini I, Felli IC, Gonnelli L, Kozminski W, Piai A, Pierattelli R, Stanek J (2012a) Speeding up sequence specific assignment of IDPs. J Biomol NMR 53:293–301
Bermel W, Bertini I, Chill J, Felli IC, Haba N, Kumar V, Pieratelli R (2012b) Exclusively heteronuclear 13C-detected amino-acid-selective NMR experiments for the study of intrinsically disordered proteins (IDP). ChemBioChem 13:2425–2432
Bermel W, Felli IC, Gonnelli L, Kozminski W, Piai A, Pieratelli R, Zawadzka-Kazimierczuk A (2013) High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR 57:353–361
Bertini I, Felli IC, Kümmerle R, Luchinat C, Pieratelli R (2004) 13C–13C NOESY: a constructive use of 13C-13C spin-diffusion. J Biomol NMR 30:245–251
Böhlen JM, Bodenhausen G (1993) Experimental aspects of chirp NMR spectroscopy. J Magn Reson Ser A 102:293–301
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Dunker AK, Silman I, Uversky VN, Sussman JL (2008) Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18:756–764
Dyson HJ, Wright PE (2001) Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol 339:258–270
Dyson HJ, Wright PE (2004) Unfolded proteins and protein folding studied by NMR. Chem Rev 104:3607–3622
Eliezer D (2009) Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol 19:23–30
Emsley L, Bodenhausen G (1992) Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J Magn Reson 97:135–148
Feng W, Rios CB, Montelione G (1996) Phase labeling of C–H and C–C spin-system topologies: application in PFG-HACANH and PFG-HACA(CO)NH triple-resonance experiments for determining backbone resonance assignments in proteins. J Biomol NMR 8:98–104
Fink AL (2005) Natively unfolded proteins. Curr Opin Struct Biol 15:35–41
Hellman M, Piirainen H, Jaakola V-P, Permi P (2014) Bridge over troubled proline: assignment of intrinsically disordered proteins using (HCA)CON(CAN)H and (HCA)N(CA)CO(N)H experiments concomitantly with HNCO and i(HCA)CO(CA)NH. J Biomol NMR 58:49–60
Hiller S, Wasmer C, Wider G, Wüthrich K (2007) Sequence-specific resonance assignment of soluble nonglobular proteins by 7D APSY-NMR spectroscopy. J Am Chem Soc 129:10823–10828
Hsu ST, Bertoncini CW, Dobson CM (2009) Use of protonless NMR spectroscopy to alleviate the loss of information resulting from exchange-broadening. J Am Chem Soc 131:7222–7223
Jensen MR, Markwick PRL, Meier S, Griesinger C, Zweckstetter M, Grzesiek S, Bernadó P, Blackledge M (2009) Quantitative determination of the conformational properties of partially folded and intrinsically disordered proteins using NMR dipolar couplings. Structure 17:1169–1185
Johnson BA, Blevins RA (1994) NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614
Kumar D, Hosur RV (2011) hNCOcanH pulse sequence and a robust protocol for rapid and unambiguous assignment of backbone (1HN, 15N and 13C′) resonances in 15N/13C-labeled proteins. Magn Reson Chem 49:575–583
Mäntylahti S, Aito O, Hellman M, Permi P (2010) HA-detected experiments for the backbone assignment of intrinsically disordered proteins. J Biomol NMR 47:171–181
Mäntylahti S, Hellman M, Permi P (2011) Extension of the HA-detection based approach: (HCA)CON(CA)H and (HCA)NCO(CA)H experiments for the main-chain assignment of intrinsically disordered proteins. J Biomol NMR 49:99–109
Marsh JA, Forman-Kay JD (2010) Sequence determinants of compaction in intrinsically disordered proteins. Biophys J 98:2383–2390
Motackova V, Novacek J, Zawadzka-Kazimierczuk A, Kazimierczuk K, Zidek L, Sanderova H, Krasny L, Kozminski W, Sklenar V (2010) Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution enhanced 5D experiments. J Biomol NMR 48:169–177
Novacek J, Haba NY, Chill JH, Zidek L, Sklenar V (2012) 4D nonuniformly sampled HCBCACON and 1J(NCα)-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J Biomol NMR 53:139–148
Pantoja-Uceda D, Santoro J (2013a) Direct correlation of consecutive C′–N groups in proteins: a method for the assignment of intrinsically disordered proteins. J Biomol NMR 57:57–63
Pantoja-Uceda D, Santoro J (2013b) A suite of amino acid residue type classification pulse sequences for 13C-detected NMR of proteins. J Magn Reson 234:190–196
Permi P, Annila A (2004) Coherence transfer in proteins. Prog NMR Spectrosc 44:97–137
Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK (2007) Intrinsic disorder and functional proteomics. Biophys J 92:1439–1456
Romero P, Obradovic Z, Li XH, Garner EC, Brown CJ, Dunker AK (2001) Sequence complexity of disordered protein. Proteins Struct Funct Genet 42:38–48
Sahu D, Bastidas M, Showalter SA (2014) Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal Biochem 449:17–25
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc 34:93–158
Shaka AJ, Keeler J, Freeman R (1983) Evaluation of a new broadband decoupling sequence: WALTZ-16. J Magn Reson 53:313–340
Shaka AJ, Barker PB, Freeman R (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson 64:547–552
Solyom Z, Schwarten M, Geist L, Konrat R, Willbold D, Brutscher B (2013) BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR 55:311–321
Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27:527–533
Tompa P (2011) Unstructural biology coming of age. Curr Opin Struct Biol 21:419–425
Tompa P (2012) Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci 37:509–516
Uversky VN, Gillespie JR, Fink AL (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Genet 41:415–427
Wen J, Wu J, Zhou P (2011) Sparsely sampled high-resolution 4-D experiments for efficient backbone resonance assignment of disordered proteins. J Magn Reson 209:94–100
Yao J, Dyson HJ, Wright PE (1997) Chemical shift dispersion and secondary structure prediction in unfolded and partially folded proteins. FEBS Lett 419:285–289
Zhang O, Forman-Kay JD, Shortle D, Kay LE (1997) Triple-resonance NOESY-based experiments with improved spectral resolution: applications to structural characterization of unfolded, partially folded and folded proteins. J Biomol NMR 9:181–200
Acknowledgments
This work was supported by project CTQ2011-22514 from the Spanish Ministerio de Economía y Competitividad. The authors thank J.L. Neira (Universidad Miguel Hernández, Alicante, Spain) and J.L. Iovanna (Centre de Recherche en Cancérologie, Marseille, France) for the Nupr1 sample.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pantoja-Uceda, D., Santoro, J. New 13C-detected experiments for the assignment of intrinsically disordered proteins. J Biomol NMR 59, 43–50 (2014). https://doi.org/10.1007/s10858-014-9827-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-014-9827-1