Abstract
Leaf senescence is an important biological process during leaf growth and development. In this study, we isolated a novel rice mutant, early senescence 3 (es3(t)), from the offspring of the wild-type rice cultivar Wuyunjing 7 after ethyl methanesulfonate mutagenesis. The es3(t) exhibited yellowing leaves, decreased chlorophyll (a and b) and carotenoid contents during the growth period, and abnormal chloroplast structure. Early leaf senescence was accompanied with decreased photosynthesis. The content of abscisic acid was increased in es3(t) mutant, which indicates that abscisic acid probably plays an important role in leaf senescence. The expression levels of senescence-associated transcription factors and senescence-associated genes were increased in the es3(t) plant, and the reactive oxygen species were accumulated in the senescence leaves of es3(t) plants. Genetic analysis demonstrated that early leaf senescence phenotype was controlled by a single recessive nuclear gene. Map-based cloning showed that ES3(t) was located on chromosome 3 with a 35-kb physical interval in the BAC AC097624 which included eight putative open reading frames. Based on these findings, we have identified a novel leaf senescence mutant which is a useful resource for revealing the molecular mechanism of early leaf senescence in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf senescence is a complex physiological process at the last stage of leaf development. It encompasses the phase from leaf maturation to death (Lim and Nam 2005; Balazadeh et al. 2011). During senescence, leaves release the mobilized nutrients and recycled them to other tissues or organs, such as seeds, storage organs, and developing leaves and flowers (Buchanan-Wollaston 1997; Thomas and Howarth 2000). However, leaves are unable to conduct photosynthesis and anabolism during leaf senescence, and the catabolism of chlorophyll becomes the dominant event. Leaf yellowing is a conveniently visible indicator of leaf senescence and mainly reflects chloroplast degradation in mesophyll cells which is the first step in senescence-associated processing (Lim et al. 2007). Early leaf senescence is an undesirable agronomic trait which diminishes crop yield by reducing the crop growth cycle. During the reproductive development and grain filling stages, early leaf senescence is closely related to the poor performance of yield-related agronomic traits (Ray et al. 1983; Bai et al. 2015; Rao et al. 2015). In contrast, appropriate timing of leaf senescence can maintain a high photosynthetic capacity and increases crop yield (Gentinetta et al. 1986; Thomas and Howarth 2000).
Leaf senescence is affected by the complex interactions between endogenous signals and environmental factors (Lim et al. 2007; Robert-Seilaniantz et al. 2011; Zhang and Zhou 2013). Abscisic acid (ABA) is a typical plant hormone with a variety of functions in stomatal aperture, seed germination and dormancy, biotic and abiotic stresses, and leaf senescence (Finkelstein and Rock 2002; Hirayama and Shinozaki 2007). The increase of endogenous ABA has been shown to coincide with leaf senescence (Gepstein and Thimann 1980; Yang et al. 2002). Generation of reactive oxygen species (ROSs) is one of the earliest responses of plant cells to abiotic stresses and senescence (Herrera-Vásquez et al. 2015). It has been demonstrated that ROS signals, especially H2O2, are involved in ABA-induced senescence of rice leaves (Hung and Kao 2004). Exogenous ABA (Biswas and Choudhuri 1980; Ray et al. 1983; Hung and Kao 2004) or H2O2 (Li et al. 2015) can accelerate the senescence of detached leaves. In addition, both ABA and ROS signalings are known to induce the expression of senescence-associated transcription factors (Zhang and Zhou 2013; Zhou et al. 2013; Wang et al. 2015) which are considered to be related to the regulation of leaf senescence.
In previous reports, several mutants with relevant phenotype affecting senescence have been mapped in rice. The lmes1 mutant exhibits spontaneous disease-like lesions in the absence of pathogen attack at the beginning of tillering stage. Malondialdehyde (MDA) was significantly increased but chlorophyll content, soluble protein content and photosynthetic rate were decreased at the booting stage, which are indicative of an early senescence phenotype. And the LMES1 gene was finally mapped to an 88-kb region with 15 ORFs (Li et al. 2014). A recessive mutation in SMS1 on chromosome 8 causes both early senescence and male sterility in rice (Yan et al. 2010). The above two genes have not yet been cloned, while some senescence-associated loci have been cloned and regulated leaf senescence through a different pathway in rice. For example, the nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence by integrating developmental cues to the jasmonic acid pathway in rice (Kong et al. 2006). The OsPSE1 may regulate premature senescence by a novel pectate lyase-mediated mechanism in rice (Wu et al. 2013). RLS1 encodes a previously uncharacterized nucleotide-binding site (NBS) containing protein with an armadillo (ARM) domain at its carboxyl terminus, and regulates rice leaf senescence through affecting chloroplast degradation (Jiao et al. 2012). Rao et al. found that the mutation of the gene encoding SCAR-like protein2 could cause drought stress and induce early senescence (Rao et al. 2015). SCAR-like protein2 is a suppressor of cAMP receptor-like protein involved in actin polymerization and panicle development in rice (Bai et al. 2015; Rao et al. 2015). PSD128/OsCDC48 encodes a putative AAA-type ATPase and is responsible for the premature senescence and death phenotype in rice. The research showed that the point mutation in PSD128/OsCDC48 made the chloroplast development impaired with significantly reduced photosynthetic ability, chlorophyll contents, root vigor, soluble protein content and increased malonaldehyde content, indicating chloroplast development and malonaldehyde content play a role in rice premature senescence (Huang et al. 2016). OsPLS1 encoding the subunit A1 of vacuolar H+-ATPase plays a causal role in premature leaf senescence through combining with ROS and salicylic acid signals (Yang et al. 2016).
In the current study, a novel leaf senescence mutant, early senescence 3 (es3(t)), was identified in rice (Oryza sativa L.). The decrease in chlorophyll contents, down-regulation of photosynthesis-associated genes and abnormal chloroplast may be responsible for the light 1000-grains weight and low seed setting rate of es3(t) mutant. The senescence-associated transcription factors and senescence-associated genes (SAGs) were up-regulated in es3(t) plants, implying that early leaf senescence occurs in es3(t) plants. The senescence-associated physiological indicators, such as endogenous ABA content and ROS signals were increased or accumulated in es3(t) plants, indicating that the ABA and ROSs probably play an important role in early leaf senescence in es3(t) mutant. A crucial link among ABA, ROSs and leaf senescence has yet to be discovered via genetic and molecular analyses in the further.
Materials and methods
Plant materials and growth conditions
The es3(t) mutant was derived from an M2 population of the japonica rice cultivar Wuyunjing 7 (WYJ7) after EMS mutagenesis. The es3(t) mutant and progenies all exhibited early leaf senescence which were stably inherited over multiple generations. The japonica cultivars WYJ7, Zhonghua 11 (ZH11) and Nipponbare (NIP) and the indica cultivars Nanjing 06 (NJ06) and 93 − 11 were used for segregating population construction. All plants were grown in the paddy fields of the China National Rice Research Institute (CNRRI) at Fuyang, Zhejiang Province, China and Lingshui, Hainan Province, China.
Pigment content measurement
The leaves (0.2 g fresh weight) from wild-type WYJ7 plant and es3(t) mutant at different growth periods, DAG15, DAG30, DAG45, DAG60, DAG75, DAG90, DAG105 (DAG, day after germination) were cut into segment, immersed in 10 ml 80 % acetone, and incubated at 26 °C in dark for 24 h. The optical density of sample solutions was measured by an ultraviolet spectrophotometer (DU800, BECKMAN, USA) at 662 nm (the maximum absorption peak of chlorophyll a), 645 nm (the maximum absorption peak of chlorophyll b) and 470 nm (the maximum absorption peak of carotenoid) light. Each sample was measured by three biological repeats. The contents of chlorophyll (Chl a and Chl b) and carotenoid (Car) in each leaf sample was calculated as follows (Arnon 1949):
where V is the volume of extraction solution (25 mL); W is the mass of rice leaf blade.
Transmission electron microscopy analysis of chloroplast ultrastructure
The middle part of flag leaves from wild-type plant (WYJ7) and es3(t) mutant at the filling stage were cut into small pieces, fixed with a phosphate buffer containing 2.5 % glutaraldehyde (pH 7.2) and vacuumed by a vacuum pumping machine for approximately 20–30 min until the specimens were immersed. The samples were then kept at 4 °C for 4 h. After fixation, they were rinsed with 0.1 M phosphate buffer (pH 7.0) and further fixed by 1 % osmium tetroxide for at least 1 h. The specimens were then dehydrated with a gradient of ethanol, embedded in small boxes and stained with lead citrate solution. The processed specimens were examined and photographed using a transmission electron microscope (Hitachi H-7650, Tokyo, Japan) (Gothandam et al. 2005; Rao et al. 2015).
Nitro blue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB) staining
Hydrogen peroxide and superoxide radicals were detected respectively using NBT and DAB according to Li et al. (2010) and Wi et al. (2010), with some modifications. Specifically, flag leaves from wild-type WYJ7 plant and es3(t) mutant at DAG105 of the filling stage were incubated in 0.05 % (w/v) NBT (Duchefa) or 0.1 % (w/v) DAB (pH 5.8, Sigma) with gentle shaking at 28 °C in dark for 12 h. The staining solution was then discarded. The chlorophyll of flag leaves was cleared by treating with 80 % (v/v) ethanol in boiling water for 10 min and then transferred to absolute ethanol. The specimens were photographed by HD Scanner.
ABA measurement
The flag leaves from wild-type WYJ7 plant and es3(t) mutant were respectively collected at DAG15 (before leaf senescence), DAG60 (senescing) and DAG105 (after leaf senescence), then immediately frozen in liquid nitrogen, and stored at −78 °C in a ultra-cold storage freezer. Powdered specimens(100 mg fresh weight) were weighed, freeze-dried for 3 h, and extracted with MilliQ water at 4 °C in dark for 16 h. Quantitative analysis of ABA was performed using the Phytodetek ABA ELISA Kit (Agdia, Inc, Elkhart, IN) according to the manufacturer’s instructions (Agrawal et al. 2001).
Lipid peroxidation and ROS-scavenging enzyme assays
Flag leaves (0.2 g fresh weight) from the wild-type WYJ7 plant and es3(t) mutant were respectively collected at DAG15 (before leaf senescence), DAG60 (senescing) and DAG105 (after leaf senescence), and then immediately frozen in liquid nitrogen. A total of 100 μl crude extract was mixed with 0.5 % (w/v) thiobarbituric acid (TBA). The mixture was then incubated at 100 °C for 15 min and centrifuged at 4 °C for 10 min. The prepared supernatants were used for measuring MDA content. The absorbance values of the supernatants were measured at 532 nm (adjust zero by distilled water) light. The MDA amounts of specimens were determined by interpolation from the standard curve (Wang et al. 2012). The powdered flag leaves were homogenized with 1 ml extraction buffer (0.1 M potassium phosphate (pH = 7.0), 1 mM EDTA, 1 % PVP (w/v), 0.1 % (v/v) Triton X-10, 1 mM PMSF) and centrifuged at 4 °C for 15 min. The prepared supernatants were used for determining superoxide dismutase (SOD) and catalase (CAT) activities which were measured respectively by a SOD analysis kit and CAT analysis kit, according to the instruction manuals (Nanjing Jiancheng Bioengineering Institute, China). The statistical analysis of the MDA, SOD and CAT contents is based on three biological replicates.
Genetic analysis and fine mapping
To determine whether a single recessive gene controls the es3(t) phenotype, the reciprocal crosses between es3(t) and the japonica cultivars NIP, WYJ7 or ZH11 were conducted. The F2 segregation populations were used for the χ2 test.
For fine mapping, the F2 segregation populations derived from the crosses between es3(t) mutant and the indica cultivars NJ06 or 93 − 11 were constructed to identify the mutated gene in es3(t) mutants. The parents and 5,013 F2 individuals were planted in the paddy field. Among them, 728 individuals with the mutant phenotype were used to map the ES3(t). PCR genotyping was carried out using a DNA bulk-pool from 42 es3(t)/NJ06 F2 individuals with the mutant phenotype, and a total of 118 SSR and sequence tagged site (STS) markers scattering among all rice chromosomes were used to determine the approximate map position of the ES3(t) locus according to Temnykh et al. (2000) and McCouch et al. (2002).
DNA extraction and molecular marker analysis
Total genomic DNA was extracted from fresh leaves using the cetyltrimethylammonium bromide-based method with minor modifications. For gene mapping, PCR-based CAPS and InDel markers were developed based on the sequence differences between the japonica rice variety Nipponbare and the indica variety 9311 (http://www.gramene.org/resources/). The primer sequences of the molecular markers are listed in Supplemental Table 2. The primers flanking the InDel polymorphisms were designed using the Primer Premier 5.0 software and tested on the parent varieties by agarose gel electrophoresis.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from the flag leaves of the wild-type WYJ7 plant and es3(t) mutant using a Total RNA Extraction Kit (Axygen, cat No, AP-MN-MS-RNA-250). Total RNA was treated with an RNase-free DNase (Promega; http://www.promega.com) and then used for complementary DNA synthesis using the ReverTra Ace qPCR-RT Kit (TOYOBA, Japan) as described by the manufacturer. Real-time PCR was performed using 2 × SYBR Green PCR Master Mix (Applied Biosystems) on the Applied Biosystems 7900HT Real-Time PCR System with three biological replicates per sample. The PCR conditions were 2 min at 50 °C, then 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The relative expression level of each transcript was compared with that of Actin1 which was selected as the internal standard for all leaf samples. The photosynthesis-associated genes (rbcL, rbcS, psaA, psbA, CAB1R, CAB2R, HEMA1, PORA and CAO) (Song et al. 2014; Tan et al. 2014), senescence-associated transcription factors (OsWRKY23, OsWRKY72 and OsNAC2), senescence-associated genes (Osl2, Osl30, Osl43, Osl57, Osl85, Osh36 and Osh69) (Wang et al. 2015), and ABA synthesis-related genes (OsNCED1, OsNCED2, OsNCED3, OsNCED4, OsNCED5) (Zhu et al. 2009) were analyzed in wild-type plant (WYJ7) and es3(t) mutant.The specific primers used for the quantification of these genes are listed in Supplemental Table 3.
Statistical analysis
All results are expressed as mean values ± standard deviation (SD) based on three biological replicates. Statistical significance was assessed using Student’s unpaired t-test. Probability values of less than 5 % were considered to be statistically significant, single asterisk (*) and double asterisk (**) represents the significant at the level of 5 and 1 %, respectively. The statistical analysis of the gene relative expression levels, physiological traits and agronomic traits in this study all follow the methods described above.
Results
Phenotypic analysis of es3(t) mutant
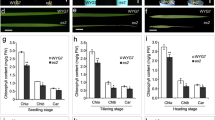
Leaf yellowing is a visible indicator of leaf senescence. There was no difference in leaf color between wild-type WYJ plant and es3(t) mutant at DAG15 before senescence (Fig. 1a). Leaf yellowing firstly appeared at DAG25 of the seedling stage and gradually became much severe along with the growth process in es3(t) (Fig. 1a). The phenotype in es3(t) mutant was most severe at DAG102 of the filling stage until harvest (Fig. 1a, b), which resulted in the defect of filling and the shrinkage of grain at the ripening stage (Fig. 1c). In addition to the obvious leaf yellowing phenotype, the es3(t) mutant exhibited a significant difference in yield-related agronomic traits, including plant height, tiller number, panicle length, 1000-grain weight and seed setting rate compared with the wild-type plant WYJ7 (Fig. 1d–h). The plant height of es3(t) was 62 ± 1.51 cm, which was 30.5 % shorter than that of the WYJ7 plant (80.9 ± 1.2 cm) (Fig. 1d). The tiller number of es3(t) was 4.25 ± 0.5, which was much fewer than that of WYJ7 (11 ± 1.8) (Fig. 1e). The panicle length of es3(t) was 10.1 ± 0.9 cm, which was shorter than that of WYJ7 (15.3 ± 0.3 cm) (Fig. 1f). The 1000-grain weight of es3(t) was 19.5 ± 0.5 g, which was lighter than that of WYJ7 (27.5 ± 0.4 g) (Fig. 1g). The seed setting rate of es3(t) was 9.7 ± 2.1 %, which was lower than that of WYJ7 (93.1 ± 2.7 %)(Fig. 1h). These observations indicate that the ES3(t) mutation affects plant height, tiller number, panicle length and seed setting rate besides leaf senescence.
Phenotype comparison of wild-type WYJ7 plant and es3(t) mutant. a The phenotype comparison of seedling and flag leaves between wild-type WYJ7 plant and es3(t) mutant at different growth period. DAG, day after germination. b The phenotype of wild-type WYJ7 plant (left) and es3(t) mutant (right) at the mature stage. c The phenotype comparison of brown rice between wild-type WYJ7 plant and es3(t) mutant. d–h Investigation of the agronomic traits including plant height (d), tiller number (e), panicle length (f), 1000-grains weight (g) and seed setting rate (h), data represent the mean ± SD of three biological replicates (Student’s t-test: *P < 0.05, **P < 0.01). Bars 2 cm in a, 10 cm in b, 3 mm in c
Genetic analysis of ES3(t)
To determine whether the es3(t) phenotype is controlled by a single recessive nuclear gene, we performed a genetic analysis of the reciprocal crosses between es3(t) and the japonica cultivars WYJ7, ZH11 or NIP. All F1 plants were as normal as wild-type WYJ7. In all three F2 segregation populations, the phenotypes of the wild-type WYJ7 plant and es3(t) mutant segregated at a ratio of 3:1 (χ2< χ2 0.05 = 3.84; P > 0.05; see Supplemental Table 1), which suggests that the phenotype of es3(t) is controlled by a single recessive nuclear gene.
Fine mapping of ES3(t)
Bulked segregant analysis (BSA) was used to produce a primary map of ES3(t). PCR genotyping was carried out using a bulk DNA pool from 42 es3(t)/NJ06 F2 individuals with the mutant phenotype, and 118 SSR and STS markers scattering on all of the rice chromosomes were used to determine the approximate map position of ES3(t). ES3(t) was primarily located on chromosome 3, closely linked to M3-1 with a genetic distance of 1.6 cM (Fig. 2a). To further fine map ES3(t), we designed some new STS and InDel markers next to M3-1 based on the sequence difference between the japonica rice variety Nipponbare and the indica variety 9311 (http://www.gramene.org/resources/). Polymorphism primers were subsequently used to screen 728 individual genotypes, and we further delimited the ES3(t) locus to an interval of 35-kb DNA region between the STS markers HK17-23 and HK17-30 on the bacterial artificial clone AC097624 (BAC1) (Fig. 2b, c). Finally, we obtained eight putative open reading frames according to the genomic annotation database RAP-DB (http://rapdb.dna.affrc.go.jp/): LOC_Os03g01140 encoding a Ecyl-activating enzyme 11, LOC_Os03g01150 encoding a Palmitoyl-protein thioesterase 1 precursor, LOC_Os03g01160 encoding a protein kinase family protein, LOC_Os03g01170 encoding a transporter family protein, LOC_Os03g01180 encoding phosphoglycerate mutase (a conserved glycolytic enzyme which has been found in the nucleoli of cancer cells), LOC_Os03g01190 encoding a dehydrogenase, LOC_Os03g01200 encoding a protein containing the SNF2 family N-terminal domain (may be involved in ATP binding), and LOC_Os03g01210 encoding an uncharacterized Cys-rich domain (Fig. 2d, e).
Fine mapping of ES3(t). a The ES3(t) locus was mapped on chromosome 3 between markers HK17-1 and M3-1. b A BAC contig spanning the ES3(t) locus. The numerals indicate the number of recombinants identified from 728 F2 mutant plants. BAC1 AC097624, BAC2 AC125411, BAC3 AC107224. c Fine mapping of ES3(t) with markers developed based on the AC097624 sequence. The ES3(t) gene was narrowed to a 35-kb genomic DNA region between STS markers HK17-23 and HK17-30. d Eight open reading frames (ORFs) as candidates for ES3(t). e Function annotation of the candidate genes
ROS and MDA accumulation were accompanied by the increased SOD activity and decreased CAT activity in es3(t) mutant
ROS generation is one of the earliest responses of plant cells during senescence (Khanna-Chopra 2012). To identify whether ROS accumulates in es3(t) plant leaves at the filling stage after senescence, we carried out the NBT staining and DAB staining. The NBT staining pattern showed the formation of blue formazan precipitates and indicates O2 − accumulation. The DAB staining pattern indicates H2O2 accumulation (Wu et al. 2016). Blue formazan precipitates of NBT staining could be observed in the leaves of es3(t), whereas the staining was minimal in the tip of wild-type leaves (Fig. 3a). The brown staining of DAB was observed in the senescence leaves of es3(t), but no such signal was detected in the leaves of the wild-type WYJ7 plant (Fig. 3b). These results suggest that ROS is accumulated in the senescence leaves of es3(t) plant.
ROS accumulation in wild-type WYJ7 and es3(t) leaves. a, b Histochemical detection of O2 − by NBT staining (a) and H2O2 by DAB staining (b) after senescence: purple formazan precipitate indicates the location of O2 − and brown precipitate indicates the location of H2O2. Flag leaves at the DAG105 of the filling stage were analyzed. c MDA content. d SOD activity. e CAT activity. For c–e, Flag leaf samples were analyzed at DAG15 (BS before senescence, seedling stage), DAG60 (S senescing, tillering stage) and DAG105(AS after senescence, filling stage); data represent the mean ± SD of three biological replicates (Student’s t-test: *P < 0.05, **P < 0.01). FW fresh weight. Bars 2 cm in a, b
Lipid peroxidation by ROS was detected by measuring the content of MDA, an end-product of oxidized lipids. Before leaf senescence at DAG15, there was no difference in leaf MDA contents between es3(t) mutant and the wild-type WYJ7 plant. The MDA content was 88.2 ± 6.3 nmol g−1 FW in es3(t) senescing flag leaves (DAG60), which was significantly higher than that of WYJ7 (47.8 ± 4.9 nmol g−1 FW). After leaf senescence at DAG105, the MDA content was 146.2 ± 8.0 nmol g−1 FW in the leaves of es3(t) mutant plants and was also much higher than that of WYJ7 (92.4 ± 5.6 nmol g−1 FW) (Fig. 3c). These results demonstrate that lipid peroxidation is significant in es3(t) mutant, which provides further evidence to reveal that ROS is accumulated in es3(t) mutant.
During oxidative stress, plants synthesize some anti-oxidative enzymes, such as SOD and CAT, to remove ROS (Miller et al. 2010). SOD catalyzes the dismutation of O2 − to produce H2O2, and CAT is the primary H2O2-scavenging enzyme. To investigate the ROS metabolic process in es3(t) plants, we detected the activities of SOD and CAT in different growth periods. There is no difference in SOD activities between es3(t) mutant and the wild-type WYJ7 plant at DAG15 before leaf senescence. The activity of SOD was 1029.1 ± 44.1 U g−1 FW in es3(t) senescing flag leaves at DAG60, which was much higher than that of WYJ7 (532.1 ± 58 U g−1 FW). We also obtained the result after leaf senescence at DAG105, with the activity of SOD of 1323.9 ± 81.6 U g−1 FW in es3(t) leaves compared with that of WYJ7 (889.6 ± 46.2 U g−1 FW) (Fig. 3d). Before leaf senescence at DAG15, there is no difference in CAT activity between es3(t) mutant and WYJ7. The activity of CAT was 26.2 ± 3.2 and 34.3 ± 2.1 U g−1 FW in es3(t) senescing flag leaves and the wild-type WYJ7 flag leaves at DAG60, respectively, and there was a significant difference in CAT activity between es3(t) and wild-type leaves. After leaf senescence at DAG105, the activity of CAT was 22.4 ± 1.9 U g−1 FW in es3(t) leaves compared with that of WYJ7 (45.9 ± 3.9 U g−1 FW) (Fig. 3e). The increase in SOD activity indicates that the es3(t) mutant may actively respond to O2 − accumulation and produce more H2O2, but the decrease in CAT activity may not be enough to scavenge the additional H2O2, leading to H2O2 accumulation in leaves and early senescence in es3(t) mutant.
Analysis of chlorophyll contents, relative expression of photosynthesis-related genes and chloroplast ultrastructure in the wild-type WYJ7 plant and es3(t) mutant
The early senescence plants which usual display yellowing might have affected photosynthesis and chlorophyll contents. To identify whether chlorophyll contents are changed in es3(t) mutant, we analyzed the chlorophyll contents in es3(t) mutant and wild-type WYJ7 plant. Kinetic analysis showed that the concentrations of chlorophyll a (Chl a), chlorophyll b (Chl b) and carotenoid in es3(t) leaves were decreased compared with those in WYJ7 at DAG30, 45, 60, 75, 90 and 105 (Fig. 4a–c).
Analysis of chlorophyll content, relative expression of photosynthesis-related genes and chloroplast ultrastructural between wild-type WYJ7 plant and es3(t) mutant. a–c Kinetic analysis of chlorophyll a content (a), chlorophyll b content (b) and carotenoid content (c). DAG, day after germination. d, e Relative expression of photosynthesis-related genes in the flag leaves of wild-type WYJ7 plant and es3(t) mutant at the seedling (d) and filling stage (e). The expression levels are relative to Actin1 mRNA. f, g Comparison of chloroplast morphology and structure between wild-type WYJ7 plant(f) and es3(t) mutant(g) by ultrastructural analysis at the filling stage. C chloroplast, N nucleus, M mitochondrionm, G grana, CW cell wall. Data represent the mean ± SD of three biological replicates (Student’s t-test: *P < 0.05, **P < 0.01)
Leaf senescence was accompanied by the decreased expression of genes related to photosynthesis (Lim et al. 2007). The expression levels of photosynthesis-associated genes (rbcL, rbcS, psaA, psbA, CAB1R, CAB2R, HEMA1, PORA and CAO) were examined in es3(t) mutant and WYJ7. The expression levels of these genes were significantly decreased in es3(t) mutant at both seedling and filling stages (Fig. 4d, e). Down-regulation of these photosynthesis-associated genes can also provide molecular evidence for early leaf senescence in es3(t) plants.
The main cellular characteristic of leaf senescence is chloroplast degradation (Lim et al. 2007; Jiao et al. 2012). To explore whether chloroplast is affected in the senescing leaves of es3(t) mutant, the ultrastructures of chloroplast were compared between es3(t) and WYJ7 at the filling stage using the transmission electron microscopy. We found that chloroplast structure was abnormal in es3(t) mutant compared with that of WYJ7 (Fig. 4f, g). In WYJ7 leaves, well-developed mesophyll cells were observed with tightly stacking lamellae in the grana of fully developed chloroplast (Fig. 4f). However, the grana lamellae stacking was absent, and the arrangement of undegraded grana was disordered in es3(t) chloroplast (Fig. 4g). These results indicate that the abnormal chloroplast is associated with leaf senescence. The decrease in chlorophyll contents, down-regulation of photosynthesis-associated genes and abnormal chloroplast may be responsible for the light 1000-grains weight and low seed setting rate of es3(t) mutant.
Identification of leaf senescence in es3(t) at the molecular level by qRT-PCR
Leaf senescence is a complicated growth and development process which is regulated by lots of genes. Previous reports indicate that the up-regulation of some transcription factors can induce leaf senescence (Zhou et al. 2013). To clarify whether the leaf senescence phenotype in es3(t) mutant was affected by senescence-associated transcription factors OsWRKY23, OsWRKY72 and OsNAC2, we performed a qRT-PCR analysis of the expression levels of these transcription factors in es3(t) mutant. The expression levels of OsWRKY23, OsWRKY72 and OsNAC2 in es3(t) leaves were increased compared with those in WYJ7 leaves at both DAG30 of the seedling stage and DAG90 of the filling stage (Fig. 5a, b). These results indicate that the expression levels of senescence-associated transcription factors were consistent with the leaf senescence in es3(t) mutant.
Identification of leaf senescence in es3(t) mutant at the molecular level by qRT-PCR. a, b Relative expression of senescence-associated transcription factors. c, d Relative expression of SAGs. Flag leaf samples of wild-type WYJ7 plant and es3(t) mutant were analyzed at the seedling (DAG30) and filling stage (DAG90). The expression levels are relative to Actin1 mRNA. Data represent the mean ± SD of three biological replicates (Student’s t-test: *P < 0.05, **P < 0.01)
We also detected the expression levels of other SAGs (Osl2, Osl30, Osl43, Osl57, Osl85, Osh36 and Osh69) using qRT-PCR. At DAG30 of the seedling stage, the mRNA of Osl2, Osl30, Osl43, Osl57, Osl85, Osh36 and Osh69 in es3(t) leaves was 1.7-, 2.2-, 22.1-, 2.1-, 14.0-,1.5- and 4.0-fold higher than those in WYJ7 leaves respectively (Fig. 5c). At DAG90 of the filling stage, their expression levels in es3(t) leaves were 3.0-, 4.4-, 15.3-, 2.9-, 18.2-, 2.3- and 7.7-fold higher than those in WYJ7 leaves, respectively (Fig. 5d). The up-regulated expression levels of SAGs further prove that early leaf senescence occurs in es3(t) plants.
ABA content and relative expressions of ABA synthesis-related genes in wild-type plant WYJ7 plant and es3(t) mutant
Previous studies indicate that ABA plays a critical role in leaf senescence (Hirayama and Shinozaki 2007; Lim et al. 2007). To identify whether the leaf senescence in es3(t) mutant is affected by ABA, we measured the ABA content at DAG15 before leaf senescence, and there was no difference in ABA contents between es3(t) and WYJ7. The content of ABA was 48.2 ± 3.6 ng g−1 FW in es3(t) senescing flag leaves at DAG60, which which was much higher than that of WYJ7 (12.9 ± 1.7 ng g−1 FW) (Fig. 6a). At DAG105 after leaf senescence, the content of ABA was 46.7 ± 4.7 ng g−1 FW in es3(t), which was significantly higher than that in WYJ7 (12 ± 3 ng g−1 FW) (Fig. 6a). In addition, the expression levels of ABA synthesis-related genes were increased in es3(t) leaves compared with those in WYJ7 leaves (Fig. 6b). These results indicate that ABA is accumulated in es3(t) plants, which probably contributes to leaf senescence in es3(t) mutant.
ABA content and relative expression level of ABA synthesis-related genes in wild-type WYJ7 plant and es3(t) mutant. a ABA content in wild-type WYJ7 and es3(t) leaves. Flag leaf samples were analyzed at DAG15 (BS, before senescence, at the seedling stage), DAG60 (S senescing, at the tillering stage) and DAG105 (AS after senescence, at the filling stage). FW fresh weight. b Relative expression level of ABA synthesis-related genes in the flag leaves of wild-type WYJ7 plant and es3(t) mutant at the DAG90 (senescing). The expression levels are relative to Actin1 mRNA. Data represent the mean ± SD of three biological replicates (Student’s t-test: *P < 0.05, **P < 0.01)
Discussion
The es3(t) is a premature leaf senescence mutant
Leaf senescence is the final stage of leaf development and critical for plants’ fitness because nutrient relocation from leaves to reproducing seeds is achieved through this process (Lim et al. 2007). Therefore, early leaf senescence (especially in functional leaves) is an extremely unfavorable phenotype for rice growth and production because of its adverse effects on photosynthetic efficiency and yield formation (Zhu et al. 2012). Isolation and characterization of mutants with altered senescence phenotype is a genetic approach to understand the molecular mechanism of leaf senescence. In this study, we identified a novel rice mutant, es3(t), with the early leaf senescence phenotype (Fig. 1). An obvious phenotypic change during leaf senescence is the gradual change of leaf color from green to yellow because of the loss of chlorophyll at the physiological level or chloroplast degradation at the cellular level (Lim et al. 2007; Jiao et al. 2012; Zhang and Zhou 2013). Besides, the expressions of photosynthesis-related genes were usually decreased during leaf senescence (Lim et al. 2007). Our results revealed that the es3(t) mutant exhibited early leaf senescence with decreased chlorophyll (a and b) and carotenoid contents compared with WYJ7 (Fig. 4a-c). Transmission electron micrography showed that chloroplast structure was abnormal in es3(t) mutant compared with that of WYJ7 (Fig. 4f, g). The decreased chlorophyll content and abnormal chloroplast might further influence the photosynthesis in es3(t) mutant (Fig. 4d, e), which was accompanied with the down-regulation of photosynthesis-related genes. At the molecular level, leaf senescence is mediated by a large number of genes, such as the senescence-associated transcription factors (OsWRKYs, OsNAC2 and AtNAP), SAGs (Osl2, Osl30, Osl43, and Osl85) and the up-regulated genes during leaf senescence (Lee et al. 2001; Guo and Gan 2006; Zhou et al. 2013). Our results showed that the expression levels of OsWRKY23, OsWRKY72, and OsNAC2 were increased in es3(t) senescing leaves compared with those in wild-type leaves (Fig. 5a, b). Besides, the expression levels of Osl2, Osl30, Osl43, Osl57, Osl85, Osh36 and Osh69 (SAGs) in es3(t) senescing leaves were significantly higher than those in wild type at both seedling and filling stages (Fig. 5c, d). Therefore, the evidence in phenotypic, physiological, cellular, and molecular levels all suggest that early leaf senescence occurs in es3(t) plants.
ES3 mutation causes early leaf senescence phenotype through affecting ABA content and ROS accumulation
From the above results, we know that early leaf senescence occurs in es3(t) mutants, but we were still not clear what caused the early senescence phenotype. Senescence is an integrated response to endogenous developmental and external environmental signals in plants. Thus, some genes involved in the response to environmental changes may regulate leaf senescence (Lim et al. 2007). Our current understanding of the relationship between environmental responses and leaf senescence is mainly based on the study of senescence response to phytohormones such as ABA, jasmonic acid, and salicylic acid which are extensively involved in the response to various stresses (Gepstein and Thimann 1980; Weaver et al. 1998; van der Graaff et al. 2006; Yang et al. 2016). These stresses can affect the hormone synthesis and/or signaling pathways to trigger the expression of stress-responsive genes, which in turn appears to affect leaf senescence. ABA is a key plant hormone mediating plants’ responses to environmental stresses. The previous study showed that ABA level was increased in senescing leaves (Biswas and Choudhuri 1980), and exogenous ABA induced the expression of several SAGs (Gepstein and Thimann 1980; Weaver et al. 1998), which is consistent with the early leaf senescence phenotypes in es3(t) mutant (Figs. 5c, d, 6a). Actually, the expression levels of the genes encoding the key enzyme in ABA biosynthesis, 9-cisepoxycarotenoid dioxygenase (NECD), is increased along with the ABA level in senescing leaves (Buchanan-Wollaston et al. 2005; van der Graaff et al. 2006). In this study, five ABA biosynthesis-related genes were up-regulated in es3(t) mutant (Fig. 6b). All these results support the role of ABA in early senescence in es3(t).
Previous reports showed that ABA induced the accumulation of H2O2 in senescing rice leaves, which in turn accelerated leaf senescence (Hung and Kao 2004). In the present study, ROSs, including O2 − and H2O2, were accumulated in the senescing leaves of es3(t) mutant (Fig. 3a, b). Meanwhile, ABA also induced the expression of antioxidant genes and enhanced the activities of antioxidative enzymes such as SOD and CAT (Hung and Kao 2003). During oxidative stress, plants synthesize anti-oxidative enzymes, such as SOD and CAT, to remove ROSs (Miller et al. 2010; Pandey and Shukla 2015). SOD catalyzes the dismutation of O2 − to produce H2O2 (Saleethong et al. 2016), and CAT is the major H2O2-scavenging enzyme. The activity of SODs in es3(t) mutant was much higher than that of the wild-type plant WYJ7 at both DAG60 of the tillering and DAG105 of the filling stage (Fig. 3d). Besides, there was no obvious difference in CAT activities between es3(t) and WYJ7 leaves at DAG15 before senescence, while the activity of CAT was decreased in es3(t) leaves compared with that of WYJ7 at both DAG60 of the tillering and DAG105 of the filling stage (Fig. 3e). Therefore, we speculate that the increase in SOD activity may actively respond to O2 − accumulation and produce more H2O2, but the decrease in CAT activity may not be enough to scavenge additional H2O2, which leads to H2O2 accumulation in leaves and early senescence in es3(t) mutant. Another explanation indicates that senescence is accelerated by the increased ABA content and may cause the increase of H2O2, since it is well known that there is an increase of ROSs during leaf senescence. MDA content analysis demonstrated that lipid peroxidation occurred in es3(t) mutant, which provided further evidence that ROSs are accumulated in es3(t) mutant. Therefore, ROSs probably play an important role in early leaf senescence in es3(t) mutant.
Moreover, genetic evidence suggests that ROSs do not trigger senescence but act as a signal activating the genetically programmed gene expression pathways, which leads to regulated cell death events (Foyer and Noctor 2005). ABA is also considered as an enhancer rather than a triggering factor for leaf senescence. Collectively, these results indicate that the ES3 mutation accelerates the leaf senescence process through combining with ROS and ABA signals. However, a crucial link among ABA, ROSs and leaf senescence has yet to be discovered via genetic and molecular analyses in the further.
Conclusion
The es3(t) mutant is characterized by early leaf senescence, decreased plant height, tiller number, panicle length and seed setting rate. Genetic analysis indicates that es3(t) is controlled by a single recessive nuclear gene. Map-based cloning reveals that the ES3(t) locus is delimited to an interval of 35-kb DNA region on chromosome 3, including eight ORFs. Physiological traits analysis suggest that the decrease in chlorophyll contents, down-regulation of photosynthesis-associated genes and abnormal chloroplast may be responsible for the light 1000-grains weight and low seed setting rate of es3(t) mutant. Moreover, the up-regulated expression levels of senescence-associated transcription factors and SAGs imply that early leaf senescence occurs in es3(t) mutant. Meanwhile, ABA and ROSs are accumulated in the senescence leaves of es3(t), indicating that ABA and ROSs probably contribute to early leaf senescence in es3(t) mutant.
References
Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol 125:1248–1257. doi:10.1104/pp.125.3.1248
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Bai JT, Zhu XD, Wang Q, Zhang J, Chen HQ, Dong GJ, Zhu L, Zheng HK, Xie QJ, Nian JQ, Chen F, Fu Y, Qian Q, Zuo JR (2015) Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol 169(2):1179–1191. doi:10.1104/pp.15.00229
Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360. doi:10.1093/mp/ssq080
Biswas AK, Choudhuri MA (1980) Mechanism of monocarpic senescence in rice. Plant Physiol 65(2):340–345. doi:10.1104/pp.65.2.340
Buchanan-Wollaston V (1997) The molecular biology of leaf senescence. J Exp Bot 48:181–199. doi:10.1093/jxb/48.2.181
Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signaling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585. doi:10.1111/j.1365-313X.2005.02399.x
Finkelstein RR, Rock CD (2002) Abscisic acid biosynthesis and response. Arabidopsis Book 1:e0058. doi:10.1199/tab.0058
Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17:1866–1875. doi:10.1105/tpc.105.033589
Gentinetta E, Ceppl D, Lepori C, Perico G, Motto M, Salamini F (1986) A major gene for delayed senescence in maize: pattern of photosynthates accumulation and inheritance. Plant Breed 97:193–203. doi:10.1111/j.1439-0523.1986.tb01053.x
Gepstein S, Thimann KV (1980) Changes in the abscisic acid content of oat leaves during senescence. Proc Natl Acad Sci USA 77:2050–2053
Gothandam KM, Kim ES, Cho HJ, Chung YY (2005) OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58(3):421–433. doi:10.1007/s11103-005-5702-5
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612. doi:10.1111/j.1365-313X.2006.02723.x
Herrera-Vásquez A, Paula Salinas P, Holuigue L (2015) Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci 6:171. doi:10.3389/fpls.2015.00171
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of th the versatile plant hormone ABA. Trends Plant Sci 12:343–351. doi:10.1016/j.tplants.2007.06.013
Huang QN, Shi YF, Zhang XB, Song LX, Feng BH, Wang HM, Xu X, Li XH, Guo D, Wu JL (2016) Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J Integr Plant Biol 58(1):12–28. doi:10.1111/jipb.12372
Hung KT, Kao CH (2003) Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J Plant Physiol 160:871–879. doi:10.1078/0176-1617-01118
Hung KT, Kao CH (2004) Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J Plant Physiol 161:1347–1357. doi:10.1016/j.jplph.2004.05.011
Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH (2012) A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol Plant 5:205–217. doi:10.1093/mp/ssr081
Khanna-Chopra R (2012) Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249:469–481. doi:10.1007/s00709-011-0308-z
Kong ZS, Li MN, Yang WQ, Xu WY, Xue YB (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141(4):1376–1388. doi:10.1104/pp.106.082941
Lee RH, Wang CH, Huang LT, Chen SC (2001) Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot 52:1117–1121. doi:10.1093/jexbot/52.358.1117
Li J, Pandeya D, Nath K, Zulfugarov IS, Yoo SC, Zhang H, Yoo JH, Cho SH, Koh HJ, Kim DS, Seo HS, Kang BC, Lee CH, Paek NC (2010) ZEBRA-NECROSIS, a thylakoidbound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. Plant J 62:713–725. doi:10.1111/j.1365-313X.2010.04183.x
Li Z, Zhang YX, Liu L, Liu QE, Bi ZZ, Yu N, Cheng SH, Cao LY (2014) Fine mapping of the lesion mimic and early senescence 1 (lmes1) in rice (Oryza sativa L.). Plant Physiol Biochem 80:300–307. doi:10.1016/j.plaphy.2014.03.031
Li ZW, Su D, Lei BT, Wang FB, Geng W, Pan G, Cheng FM (2015) Transcriptional profile of genes involved in ascorbate glutathione cycle in senescing leaves for an early senescence leaf (esl) rice mutant. J Plant Physiol 176:1–15. doi:10.1016/j.jplph.2014.09.020
Lim PO, Nam HG (2005) The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr Top Dev Biol 67:49–83. doi:10.1016/S0070-2153(05)67002-0
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136. doi:10.1146/annurev.arplant.57.032905.105316
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res 9:199–207. doi:10.1093/dnares/9.6.199
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467. doi:10.1111/j.1365-3040.2009.02041.x
Pandey V, Shukla A (2015) Acclimation and tolerance strategies of rice under drought stress. Rice Sci 22(4):147–161. doi:10.1016/S1672-6308(14)60289-4
Rao YC, Yang YL, Xu J, Li XJ, Leng YJ, Dai LP, Huang LC, Shao GS, Ren DY, Hu J, Guo LB, Pan JW, Zeng DL (2015) EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol 169(2):1225–1239. doi:10.1104/pp.15.00991
Ray S, Mondal WA, Choudhuri MA (1983) Regulation of leaf senescence, grain-filling and yield of rice by kinetin and abscisic acid. Physiol Plant 59:343–346. doi:10.1111/j.1399-3054.1983.tb04212.x
Robert-Seilaniantz A, Grant M, Jones JD (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49:317–343. doi:10.1146/annurev-phyto-073009-114447
Saleethong P, Roytrakul S, Kong-Ngern K, Theerakulpisut P (2016) Differential proteins expressed in rice leaves and grains in response to salinity and exogenous spermidine treatments. Rice Sci 23(1):9–21. doi:10.1016/j.rsci.2016.01.002
Song J, Wei XJ, Shao GN, Sheng ZH, Chen DB, Liu CL, Jiao GA, Xie LH, Tang SQ, Hu PS (2014) The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein in needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol 84(3):301–304. doi:10.1007/s11103-013-0134-0
Tan JJ, Tan ZH, Wu FQ, Sheng PK, Heng YQ, Wang XH, Ren YL, Wang JL, Guo XP, Zhang X, Cheng ZJ, Jiang L, Liu XM, Wang HY, Wan JM (2014) A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol Plant 7(8):1329–1349. doi:10.1093/mp/ssu054
Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice. Theor Appl Genet 100:697–712. doi:10.1007/s001220051342
Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51:329–337. doi:10.1093/jexbot/51
van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792. doi:10.1104/pp.106.079293
Wang X, Fang G, Li Y, Ding M, Gong HY, Li YS (2012) Differential antioxidant responses to cold stress in cell suspension cultures of two subspecies of rice. Plant Cell Tissue Org Cult 113:353–361. doi:10.1007/s11240-012-0273-z
Wang ZH, Wang Y, Hong X, Hu DH, Liu CX, Yang J, Li Y, Huang YQ, Feng YQ, Gong HY, Li Y, Fang G, Tang HR, Li YS (2015) Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J Exp Bot 66(3):973–987. doi:10.1093/jxb/eru456
Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37:455–469. doi:10.1023/A:1005934428906
Wi SJ, Jang SJ, Park KY (2010) Inhibition of biphasic ethylene production enhances tolerance to abiotic stress by reducing the accumulation of reactive oxygen species in Nicotiana tabacum. Mol Cells 30:37–49. doi:10.1007/s10059-010-0086-z
Wu HB, Wang B, Chen YL, Liu YG, Chen LT (2013) Characterization and fine mapping of the rice premature senescence mutant ospse1. Theor Appl Genet 126(7):1897–1907. doi:10.1007/s00122-013-2104-y
Wu LW, Ren DY, Hu SK, Li GM, Dong GJ, Jiang L, Hu XM, Ye WJ, Cui YT, Zhu L, Hu J, Zhang GH, Gao ZY, Zeng DL, Qian Q, Guo LB (2016) Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol 171:1085–1098. doi:10.1104/pp.15.01898
Yan WY, Ye SH, Jin QS, Zeng LJ, Peng Y, Yan DW, Yang WB, Yang DL, He ZH, Dong YJ, Zhang XM (2010) Characterization and mapping of a novel mutant sms1 (senescence and male sterility 1) in rice. J Genet Genomics 37:47–55. doi:10.1016/S1673-8527(09)60024-2
Yang JC, Zhang JH, Wang ZQ, Zhu QS, Liu LJ (2002) Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta 215:645–652. doi:10.1007/s00425-002-0789-2
Yang X, Gong P, Li K, Huang F, Cheng F, Pan G (2016) A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. J Exp Bot. doi:10.1093/jxb/erw109 [Epub ahead of print]
Zhang H, Zhou C (2013) Signal transduction in leaf senescence. Plant Mol Biol 82:539–545. doi:10.1007/s11103-012-9980-4
Zhou QY, Yu Q, Wang ZQ, Pan YF, Lv WT, Zhu LL, Chen RZ, He GC (2013) Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ 36:1476–1489. doi:10.1111/pce
Zhu GH, Ye NH, Zhang JH (2009) Glucose-Induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol 50(3):644–651. doi:10.1093/pcp/pcp022
Zhu LF, Yu SM, Jin QY (2012) Effects of aerated irrigation on leaf senescence at late growth stage and grain yield of rice. Rice Sci 19: 44–48. doi:10.1016/S1672-6308(12)60019-5
Acknowledgments
This study was funded by National Natural Science Foundation of China (31501279), China Postdoctoral Science Foundation (2015 M570181) and Shenzhen Scientific and Technological Program (JCYJ20140504111101999, JCYJ20150630165133402).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Yan Su, Shikai Hu and Bin Zhang have equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Su, Y., Hu, S., Zhang, B. et al. Characterization and fine mapping of a new early leaf senescence mutant es3(t) in rice. Plant Growth Regul 81, 419–431 (2017). https://doi.org/10.1007/s10725-016-0219-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0219-2