Abstract
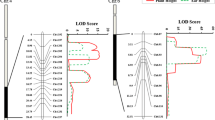
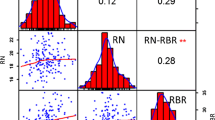
Plant height (PH) and main shoot length (MSL) are multi-allelic quantitative traits, that exert profound influences on crop productivity. The identification of QTLs responsible for PH and MSL is important for enhancing crop yield potential. To achieve this, we constructed a genetic linkage map using 192 SSRs in combination with 94 BCILs derived from JS1 and JS2 somatic hybrids. This comprehensive map spanned a total length of 1420.69 cM, expanding into 12 linkage groups. For PH, we identified a total of seven and one QTLs across two successive crop seasons. Interestingly, the minor QTLs identified during the first season exhibited relatively modest phenotypic effects (2.43–7.33%) while a dominant QTL (12.88% PVE) was identified in the subsequent crop season. All these QTLs displayed positive and negative additive effects, often accompanied by exceptionally high dominance effects (≤ 39.06). For MSL, we detected three QTLs during the first crop season, characterized by LOD values of 3.0063, 3.3156, and 8.9608, explaining 5.61%, 6.17%, and 16.27% PVE, respectively. Subsequently, two QTLs were identified on LG-2 with LOD values of 2.5869 (13.79% PVE) and 3.5740 (7.93% PVE), revealing negative additive effects. In the QTL regions for PH and MSL, we discovered a total of 20 genes, with eight and twelve genes associated with auxin and Brassinosteroids genes, displaying essential roles in regulating shoot height. In this pioneering study, we present the first-ever report detailing the identification of PH and MSL genes contributed by S. alba creating linkage drags in advanced BCILs and providing an opportunity for Brassica crop improvement.





Similar content being viewed by others
Data availability
All the data generated during the current research work is available with the manuscript and its supplementary files.
Abbreviations
- LOD:
-
logarithm of the odds
- PVE:
-
Phenotypic variance
- Add:
-
Additive Effect
- Dom:
-
Dominance Effect
- CI:
-
Confidence interval
References
Addinsoft (2023) XLSTAT statistical and data analysis solution. New York, USA. https://www.xlstat.com/en
Agrawal N, Gupta M, Atri C, Akhatar J, Kumar S, Heslop-Harrison P, Banga SS (2021) Anchoring alien chromosome segment substitutions bearing gene(s) for resistance to mustard aphid in Brassica juncea-B. fruticulosa introgression lines and their possible disruption through gamma irradiation. Theor Appl Genet 134:3209–3224. https://doi.org/10.1007/s00122-021-03886-z
Asgher M, Khan MI, Anjum NA, Khan NA (2015) Minimising toxicity of cadmium in plants-role of plant growth regulators. Protoplasma 252:399–413. https://doi.org/10.1007/s00709-014-0710-4
Atri C, Akhatar J, Gupta M et al (2019) Molecular and genetic analysis of defensive responses of Brassica juncea – B. fruticulosa introgression lines to Sclerotinia Infection. Sci Rep 9:17089. https://doi.org/10.1038/s41598-019-53444-3
Bohra A, Kilian B, Sivasankar S, Caccamo M, Mba C, McCouch SR, Varshney RK (2022) Reap the crop wild relatives for breeding future crops. Trends Biotechnol 40(4):412–431. https://doi.org/10.1016/j.tibtech.2021.08.009
Cai G, Yang Q, Chen H, Yang Q, Zhang C, Fan C et al (2016) Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci Rep 6:21625. https://doi.org/10.1038/srep21625
Chaiwanon J, Wang ZY (2015) Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25:1031–1042. https://doi.org/10.1016/j.cub.2015.02.046
Chen BY, Xu K, Li J, Li F, Qiao JW, Li H et al (2014) Evaluation of yield and agronomic traits and their genetic variation in 488 global collections of Brassica napus L. Genet Resour Crop Evol 61:979–999. https://doi.org/10.1007/s10722-014-0091-8
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Biol 49:427–451. https://doi.org/10.1146/annurev.arplant.49.1.427
Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111:671–678
Ding G, Zhao Z, Liao Y, Hu Y, Shi L, Long Y et al (2012) Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Ann Bot 109:747–759. https://doi.org/10.1093/aob/mcr323
Dong Z, Alam MK, Xie M, Yang L, Liu J, Helal MMU, Huang J, Cheng X, Liu Y, Tong C, Zhao C, Liu S (2021) Mapping of a major QTL controlling plant height using a high-density genetic map and QTL-seq methods based on whole-genome resequencing in Brassica napus. G3 (Bethesda) 11(7):jkb118. https://doi.org/10.1093/g3journal/jkab118
Evenson RE, Gollin D (2003) Assessing the impact of the green revolution, 1960 to 2000. Science 300:758–762. https://doi.org/10.1126/science.1078710
Gaikwad K, Kirti PB, Sharma A, Prakash S, Chopra VL (1996) Cytogenetical and molecular investigations on somatic hybrids of Sinapis alba and Brassica juncea and their backcross progeny. Plant Breed 115(6):480–483. https://doi.org/10.1111/j.1439-0523.1996.Tb00961.x
Garg H, Li H, Sivasithamparam K, Kuo J, Barbetti MJ (2010) The Infection processes of Sclerotiniasclerotiorum in cotyledon tissue of a resistant and a susceptible genotype of Brassica napus. Ann Bot 106(6):897–908. https://doi.org/10.1093/aob/mcq196
Groot SPC, Karssen CM (1987) Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta 171:525–531. https://doi.org/10.1007/BF00392302
Gudi S, Atri C, Goyal A et al (2020) Physical mapping of introgressed chromosome fragment carrying the fertility restoring (Rfo) gene for Ogura CMS in Brassica juncea L. Czern & Coss. Theo Appl Genet 133:2949–2959. https://doi.org/10.1007/s00122-020-03648-3
Guilfoyle TJ, Ulmasov T, Hagen G (1998) The ARF family of transcription factors and their role in plant hormone responsive transcription. Cell Mol Life Sci 54:619–627. https://doi.org/10.1007/s000180050190
Kaur G, Singh VV, Singh KH, Priyamedha Rialch I, Gupta M et al (2022) Classical genetics and traditional breeding in Brassica juncea. In: Kole C, Mohapatra T (eds) The Brassica juncea Genome: Compendium of plant genomes. Springer, Cham, pp 85–113. https://doi.org/10.1007/978-3-030-91507-0_4
Kirti PB, Mohapatra T, Khanna H, Prakash S, Chopra VL (1995) Diplotaxis catholica + Brassica juncea somatic hybrids: molecular and cytogenetic characterization. Plant Cell Rep 14:593–597
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Kumari P, Bhat SR (2019) Allohexaploid (H2) (IC0626000 INGR18031), an Indian mustard (Brassica juncea) germplasm with heat tolerance and Resistant to Alternaria brassicae. Indian J Plant Genet Res 32(3):439
Kumari P, Singh KP (2019) Characterization of stable somatic hybrids of Sinapis alba and Brassica juncea for Alternaria blight, Sclerotinia sclerotiurum resistance and heat tolerance. Indian Res J Ext Edu 19(2&3):99–103
Kumari P, Bhat SR (2021) Allohexaploid (H1)(IC0628060; INGR19102), an allohexaploid (Brassica juncea + Sinapis alba) germplasm resistant to Alternariabrassicae, Sclerotiniasclerotiorum, and tolerant to temperature. Indian J Plant Genet Resour 34(1):160–161
Kumari P, Bist DS, Bhat SR (2018) Stable fertile somatic hybrids between Sinapis alba and Brassica juncea showing resistance to Alternaria brassicae and heat stress. Plant Cell Tissue Organ Cult 133(1):77–86. https://doi.org/10.1007/s11240-017-1362-9
Kumari P, Singh KP, Rai PK (2020a) Draft genome of multiple resistance donor plant Sinapisalba: an insight into SSRs, annotations and phylogenetics. PLOS ONE. https://doi.org/10.1371/journal.pone
Kumari P, Singh KP, Bisht D, Kumar S (2020b) Somatic hybrids of Sinapis alba + Brassica juncea: study of backcross progenies for morphological variations, chromosome constitution, and reaction to Alternaria brassicae. Euphytica 216:93. https://doi.org/10.1007/s10681-020-02629-3
Kumari P, Singh KP, Kumar S, Yadava DK (2020c) Development of a yellow-seeded stable allohexaploid brassica through inter-generic somatic hybridization with a high degree of fertility and resistance to Sclerotinia Sclerotiorum. Front Plant Sci 11:575591. https://doi.org/10.3389/fpls.2020.575591
Kumari P, Singh KP, Rai PK (2023) Identification of new resistance source for Sclerotinia stem rot in backcross population of B. juncea + S. alba allohexaploids: key to manage disease through host resistance. Euphytica 219:76. https://doi.org/10.1007/s10681-023-03208-y
Landry C, Hart ID, Ranz J (2007) Genome clashes in hybrids: insights from gene expression. Heredity 99:483–493
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272:398–401
Li AM, Wei CX, Jiang JJ, Zhang YT, Snowdon RJ, Wang Y (2009) Phenotypic variation in the progeny of somatic hybrids between Brassica napus and Sinapis alba. Euphytica 170:289–296. https://doi.org/10.1007/s10681-009-9979-3
Li X, Chen L, Hong M, Zhang Y, Zu F, Wen J et al (2012) A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed Coat in Brassica rapa. PLoS ONE 7(9):e44145. https://doi.org/10.1371/journal.pone.0044145
Li X, Ramchiary N, Dhandapani V, Choi SR, Hur Y, Nou IS, Yoon MK, Lim YP (2013) Quantitative trait loci mapping in Brassica rapa revealed the structural and functional conservation of genetic loci governing morphological and yield component traits in the A, B, and C subgenomes of Brassica species. DNA Res 20(1):1–16. https://doi.org/10.1093/dnares/dss029
Liu C, Wang J, Huang T, Wang F, Yuan F, Cheng X et al (2010) A missense mutation in the VHYNP motif of a DELLA protein causes a semi- dwarf mutant phenotype in Brassica napus. Theor Appl Genet 121:249–258. https://doi.org/10.1007/s00122-010-1306-9
Liu Z, Wang Y, Shen GYW, Hao S, Liu B (2004) Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia. Plant Mol Biol 54:571–582
Mauricio R (2001) Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat Rev Genet 2:370–381
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3(3):269–283. https://doi.org/10.1016/j.cj.2015.01.001
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE et al (1999) Green revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261. https://doi.org/10.1038/22307
Qi L (2014) QTL analysis for the traits Associated with Plant Architecture and Silique in Brassica napus L. Huazhong Agricultural University, Doctor, wuhan
Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R et al (2006) A comparative linkage map of oilseed Rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114:67–80. https://doi.org/10.1007/s00122-006-0411-2
Quijada PA, Udall JA, Lambert B, Osborn TC (2006) Identification of genomic regions from winter germplasm that affect seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L). Theor Appl Genet 113:549–561
Rana K, Atri C, Gupta M, Akhatar J, Sandhu PS et al (2017) Mapping resistance responses to Sclerotinia infestation in introgression lines of Brassica juncea carrying genomic segments from wild Brassicaceae B. fruticulosa. Sci Rep 7:5904. https://doi.org/10.1038/s41598-017-05992-9
Rana K, Atri C, Akhatar J, Kaur R, Goyal A, Singh MP et al (2019) Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea-Erucastrum cardaminoides introgression lines. Front Plant Sci 10:1015. https://doi.org/10.3389/fpls.2019.01015
Saini S, Sharma I, Pati PK (2015) Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front Plant Sci 6:950. https://doi.org/10.3389/fpls.2015.00950
Sethi M, Saini DK, Devi V, Kaur C, Singh MP, Singh J, Pruthi G, Kaur A, Singh A, Chaudhary DP (2023) Unravelling the genetic framework associated with grain quality and yield-related traits in maize (Zea mays L). Front Genet 7 14:1248697. https://doi.org/10.3389/fgene.2023.1248697
Shen Y, Xiang Y, Xu E, Ge X, Li Z (2018) Major co-localized QTL for plant height, branch ınitiation height, stem diameter, and flowering time in an alien ıntrogression derived Brassica napus DH population. Front Plant Sci 9:390. https://doi.org/10.3389/fpls.2018.00390
Singh RK, Chaudhary BD (1977) Biometrical methods in quantitative genetic analysis. Kalyani publishers, New Delhi (304)
Singh KP, Kumari P, Rai PK (2021a) Current status of the disease-resistant gene(s)/QTLs, and strategies for improvement in Brassica juncea. Front Plant Sci 12:617405. https://doi.org/10.3389/fpls.2021.617405
Singh KP, Kumari P, Yadava DK (2021b) Introgression and QTL mapping conferring resistance for Alternaria brassicae in the backcross progeny of Sinapis alba + Brassica juncea somatic hybrids. Plant Cell Rep 40:2409–2419. https://doi.org/10.1007/s00299-021-02785-3
Singh KP, Kumari P, Yadava DK (2022a) Development of de-novo transcriptome assembly and SSRs in Allohexaploid Brassica with functional annotations and identification of heat-shock proteins for thermotolerance. Front Genet 13:958217. https://doi.org/10.3389/fgene.2022.958217
Singh KP, Kumari P, Raipuria RK, Rai PK (2022b) Development of genome-specific SSR markers for the identification of introgressed segments of Sinapis alba carrying genes for blight resistance in the Brassica juncea background. 3Biotechnology 12:332. https://doi.org/10.1007/s13205-022-03402-0
Singh KP, Kumari P, Rai PK (2023) Phenotypic characterization and resistance response to Sclerotinia sclerotiorum of backcross lines developed from stable allohexaploids of Sinapis alba + Brassica juncea. Euphytica 219:34. https://doi.org/10.1007/s10681-023-03160-x
Song KM, Slocum MK, Osborn TC (1995) Molecular marker analysis of genes controlling morphological variation in Brassica rapa (syn. campestris). Theor Appl Genet 90:1–10
Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85:171–182
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203. https://doi.org/10.1007/BF00223376
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543. https://doi.org/10.1105/tpc.008417
Udall JA, Quijada PA, Lambert B, Osborn TC (2006) Identification of alleles from unadapted germplasm affecting seed yield and other quantitative traits in hybrid spring oilseed Brassica napus L. Theor Appl Genet 113:597–609
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19:309–319. https://doi.org/10.1046/j.1365-313X.1999.00538.x
Wang YP, Sonntag K, Rudloff E, Chen JM (2005) Intergeneric somatic hybridization between Brassica napus L. and Sinapis alba L. J Integr Plant Biol 47(1):84–91. https://doi.org/10.1111/j.1744-7909.2005.00009.x
Wang J, Gao Y, Kong Y, Jiang J, Li A, Zhang Y, Wang Y (2014) Abortive process of a novel rapeseed cytoplasmic male sterility line derived from somatic hybrids between Brassica napus and Sinapis alba. J Integr Agric 13(4):741–748. https://doi.org/10.1016/S2095-3119(13)60584-5
Wang J, Jing L, Jian H, Qu C, Chen L, Li J et al (2015) Quantitative trait loci mapping for plant height, the first branch height, and branch number and possible candidate genes screening in Brassica napus L. Acta Agron Sin 41:1027–1038. https://doi.org/10.3724/SP.J.1006.2015.01027
Wang X, Wang H, Long Y, Liu L, Zhao Y, Tian J et al (2015) Dynamic and comparative QTL analysis for plant height in different developmental stages of Brassica napus L. Theor Appl Genet 128:1175–1192. https://doi.org/10.1007/s00122-015-2498-9
Wang Y, He J, Yang L, Wang Y, Chen W, Wan S, Chu P, Guan R (2016) Fine mapping of a major locus controlling plant height using a high-density single-nucleotide polymorphism map in Brassica napus. Theor Appl Genet 129(8):1479–1491. https://doi.org/10.1007/s00122-016-2718-y
Wang Y, Zhao J, Lu W, Deng D (2017) Gibberellin in plant height control: old player, new story. Plant Cell Rep 36:391–398. https://doi.org/10.1007/s00299-017-2104-5
Whitney KD, Gabler CA (2008) Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Divers Distrib 14:569–580
Yi B, Chen W, Ma C, Fu T, Tu J (2006) Mapping of quantitative trait loci for yield and yield components in Brassica napus L. Acta Agron Sin 32:676–682
Yu SC, Wang YJ, Zheng XY (2003) Mapping and analysis QTL controlling some morphological traits in Chinese cabbage (Brassica campestris L. ssp. pekinensis). Yi Chuanxuebao =. Acta Genet Sin 30(12):1153–1160
Zhang F, Liu Y, Cheng X, Tong C, Dong C, Tang M et al (2014) QTL mapping of plant height using high density SNP markers in Brassica napus. Chin J Oil Crop Sci 36:695–700. https://doi.org/10.7505/j.issn.1007-9084.2014.06.001
Zhu T, Liu B, Liu N, Xu J, Song X, Li S, Sui S (2022) Gibberellin related genes regulate dwarfing mechanism in winter sweet. Front Plant Sci 13:1010896. https://doi.org/10.3389/fpls.2022.1010896
Funding
(1) Preetesh Kumari: Acknowledges the Department of Science and Technology (DST), Ministry of Science and Technology, New Delhi, Govt. of India, India for research grant [File No. SR/WOS-A/LS-373/2018(G)]; (2) Kaushal Pratap Singh received a fellowship (Research Associate) grant (File No. 09/1247(0001)/2019-EMR-I) from the HRDG-Council of Scientific and Industrial Research (HRDG-CSIR), New Delhi.
Author information
Authors and Affiliations
Contributions
PK: developed the somatic hybrids and their backcross progenies, conceived the study, fund acquisition, prepared the first draft and finalization of the manuscript; KPS conducted field trials for both years, phenotyping, field data analysis, genotyping, QTL mapping, editing, and finalization of the manuscript; PKR: Provided laboratory and field facilities to conduct this study, read the manuscript. All authors read the final version of the manuscript and approved it for submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10681_2023_3271_MOESM2_ESM.xls
Details of SSR markers with SSR locations, left and right marker sequences, Tm, and product size used in the present study for QTL mapping (XLS 105.5 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumari, P., Singh, K.P. & Rai, P.K. Deciphering the genetic architecture of plant height and main shoot length in backcross introgression lines of Sinapis alba + Brassica juncea allohexaploids. Euphytica 220, 3 (2024). https://doi.org/10.1007/s10681-023-03271-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03271-5