Abstract
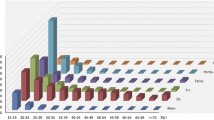
A large set of potential microsatellites markers covering the entire genome are required for genetic diversity, mapping and functional genomics studies. In this study, a total of 37,572 microsatellites with frequency of 83.25 microsatellites per Mb were mined in the recently sequenced Fagopyrum tataricum. Among these 37,572 microsatellites, most of the microsatellites were di-nucleotide (26,196, 69.72%) and AT/TA repeat motifs were the most abundant (24,742, 65.85%). A total of 26,549 microsatellite markers were designed from the flanking sequences of 37,572 microsatellites and 2643 (9.96%) were genic microsatellite markers. All of these newly-developed microsatellite markers were physically anchored to the eight chromosomes of F. tataricum and the first and high-density physical map was constructed with an average marker density of 58.82 markers per Mb. Amplification efficiency and experimental evaluation of the newly-developed microsatellite markers assessed with 440 markers randomly selected in F. tataricum, showed that these microsatellite markers are highly effective and valuable. The large number of new microsatellite markers and their location on the physical map provided a valuable resource for studying diversity, constructing genetic maps, functional gene mapping, QTL exploration and molecular breeding in Fagopyrum species.




Similar content being viewed by others
References
Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep 27(4):617–631
Blanca J, Cañizares J, Roig C, Ziarsolo P, Nuez F, Picó B (2011) Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae). BMC Genom 12(1):104
Bonafaccia G, Marocchini M, Kreft I (2003) Composition and technological properties of the flour and bran from common and Tartary buckwheat. Food Chem 80(1):9–15
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD et al (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom 11(1):569
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCOUCH SR et al (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100(5):713–722
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363(1491):557–572
Da Maia LC, Palmieri DA, De Souza VQ, Kopp MM, de Carvalho FIF, Costa de Oliveira A (2008) SSR locator: tool for simple sequence repeat discovery integrated with primer design and PCR simulation. Int J Plant Genom 2008:412696
Fabjan N, Rode J, Košir IJ, Wang Z, Zhang Z, Kreft I (2003) Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J Agric Food Chem 51(22):6452–6455
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113(3):163–185
Gupta S, Kumari K, Sahu PP, Vidapu S, Prasad M (2012) Sequence-based novel genomic microsatellite markers for robust genotyping purposes in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Cell Rep 31(2):323–337
Hou SY, Sun ZX, Linghu B, Xu DM, Wu B, Zhang B, Wang XC, Han YH, Zhang LJ, Qiao ZJ, Li HY (2016) Genetic diversity of buckwheat cultivars (Fagopyrum tartaricum Gaertn.) assessed with SSR markers developed from genome survey sequences. Plant Mol Biol Rep 34(1):233–241
Iwata H, Imon K, Tsumura Y, Ohsawa R (2005) Genetic diversity among Japanese indigenous common buckwheat (Fagopyrum esculentum) cultivars as determined from amplified fragment length polymorphism and simple sequence repeat markers and quantitative agronomic traits. Genome 48(3):367–377
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177(3):309–334
Kishore G, Pandey A, Dobhal R, Gupta S (2013) Population genetic study of Fagopyrum tataricum from Western Himalaya using ISSR markers. Biochem Genet 51(9–10):750–765
Konishi T, Ohnishi O (2006) A linkage map for common buckwheat based on microsatellite and AFLP markers. Fagopyrum 23(2):1–6
Lagercrantz U, Ellegren H, Andersson L (1993) The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res 21(5):1111–1115
Li YQ, Shi TL, Zhang ZW (2007) Development of microsatellite markers from Tartary buckwheat. Biotechnol Lett 29(5):823–827
Ma KH, Kim NS, Lee GA, Lee SY, Lee JK, Yi JY et al (2009) Development of SSR markers for studies of diversity in the genus Fagopyrum. Theor Appl Genet 119(7):1247–1254
Morgante M, Olivieri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3(1):175–182
Pan SJ, Chen QF (2010) Genetic mapping of common buckwheat using DNA, protein and morphological markers. Hereditas 147(1):27–33
Pandey G, Misra G, Kumari K, Gupta S, Parida SK, Chattopadhyay D, Prasad M (2013) Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res 20(2):197–207
Parida SK, Kumar KAR, Dalal V, Singh NK, Mohapatra T (2006) Unigene derived microsatellite markers for the cereal genomes. Theor Appl Genet 112(5):808–817
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1(7):215–222
Puranik S, Sahu PP, Mandal SN, Parida SK, Prasad M (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PLoS ONE 8(5):e64594
Rohlf FJ (1992) NTSYS-PC: numerical taxonomy and multivariate analysis system version 2.02 k. State University of New York, Stony Brook, NY
Schlötterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nucleic Acids Res 20(2):211–215
Sharma T, Jana S (2002) Species relationships in Fagopyrum revealed by PCR-based DNA fingerprinting. Theor Appl Genet 105(2–3):306–312
Sharma PC, Grover A, Kahl G (2007) Mining microsatellites in eukaryotic genomes. Trends Biotechnol 25(11):490–498
Shi J, Huang S, Zhan J, Yu J, Wang X, Hua W et al (2013) Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res 21(1):53–68
Shi T, Li R, Chen Q, Li Y, Pan F, Chen Q (2017) De novo sequencing of seed transcriptome and development of genic-SSR markers in common buckwheat (Fagopyrum esculentum). Mol Breed 37(12):147
Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN et al (2011) Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS ONE 6(6):e21298
Suzuki T, Morishita T, Mukasa Y, Takigawa S, Yokota S, Ishiguro K, Noda T (2014a) Breeding of ‘Manten-Kirari’, a non-bitter and trace-rutinosidase variety of Tartary buckwheat (Fagopyrum tataricum Gaertn.). Breed Sci 64(4):344–350
Suzuki T, Morishita T, Mukasa Y, Takigawa S, Yokota S, Ishiguro K, Noda T (2014b) Discovery and genetic analysis of non-bitter Tartary buckwheat (Fagopyrum tataricum Gaertn.) with trace-rutinosidase activity. Breed Sci 64(4):339–343
Tang Y, Ding MQ, Tang YX, Wu YM, Shao JR, Zhou ML (2016) Germplasm resources of buckwheat in China. In: Zhou M, Kreft I, Woo SH, Chrungoo NK, Wieslander G (eds) Molecular breeding and nutritional aspects of buckwheat. Academic Press, The Netherlands, pp 13–20
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12(10):4127–4138
Tsuji K, Ohnishi O (2001) Phylogenetic relationships among wild and cultivated Tartary buckwheat (Fagopyrum tataricum Gaert.) populations revealed by AFLP analyses. Genes Genet Syst 76(1):47–52
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Wang Y, Campbell CG (2007) Tartary buckwheat breeding (Fagopyrum tataricum L. Gaertn.) through hybridization with its Rice-Tartary type. Euphytica 156(3):399–405
Wang Q, Fang L, Chen J, Hu Y, Si Z, Wang S et al (2015) Genome-wide mining, characterization, and development of microsatellite markers in Gossypium species. Sci Rep 5:10638
Yabe S, Hara T, Ueno M, Enoki H, Kimura T, Nishimura S et al (2014) Rapid genotyping with DNA micro-arrays for high-density linkage mapping and QTL mapping in common buckwheat (Fagopyrum esculentum Moench). Breed Sci 64(4):291–299
Yasodha R (2011) Characterization of microsatellites in the tribe Bambuseae. Gene Conserve 10(39):51–64
Yasui Y, Wang Y, Ohnishi O, Campbell CG (2004) Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self-pollinated relative Fagopyrum homotropicum. Genome 47(2):345–351
Yasui Y, Hirakawa H, Ueno M, Matsui K, Katsube-Tanaka T, Yang SJ et al (2016) Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res 23(3):215–224
Zhang ZS, Xiao YH, Luo M, Li XB, Luo XY, Hou L et al (2005) Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.). Euphytica 144(1–2):91–99
Zhang HB, Li Y, Wang B, Chee PW (2008) Recent advances in cotton genomics. Int J Plant Genom 2008:742304
Zhang L, Li X, Ma B, Gao Q, Du H, Han Y et al (2017) The Tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol Plant 10(9):1224–1237
Zhao H, Yang L, Peng Z, Sun H, Yue X, Lou Y et al (2015) Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci Rep 5:8018
Zhu H, Senalik D, McCown BH, Zeldin EL, Speers J, Hyman J et al (2012) Mining and validation of pyrosequenced simple sequence repeats (SSRs) from American cranberry (Vaccinium macrocarpon Ait.). Theor Appl Genet 124(1):87–96
Zou C, Lu C, Zhang Y, Song G (2012) Distribution and characterization of simple sequence repeats in Gossypium raimondii genome. Bioinformation 8(17):801
Acknowledgements
This study was financially supported by Chinese Postdoctoral Science Foundation (2017M622944), Chongqing Postdoctoral Science Foundation (No. Xm2017176), and Fundamental Research Funds for the Central Universities (No. XDJK2018C051).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
The physical map of the 26,549 microsatellite markers of F. tataricum. The eight chromosomes of Tartary buckwheat are arranged from left to right. The background is green color and the blue lines are the positions of microsatellite markers. (PNG 1796 kb)
Figure S2
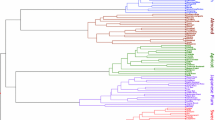
Dendrograms of 12 Fagopyrum varieties based on the 894 polymorphic loci from the 440 SSR markers (A), 748 loci form inter-genic SSR markers (B), 146 loci from intra-genic SSR markers (C). (PNG 1032 kb)
Rights and permissions
About this article
Cite this article
Fang, X., Huang, K., Nie, J. et al. Genome-wide mining, characterization, and development of microsatellite markers in Tartary buckwheat (Fagopyrum tataricum Garetn.). Euphytica 215, 183 (2019). https://doi.org/10.1007/s10681-019-2502-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2502-6