Abstract
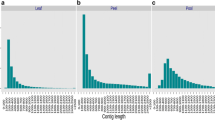
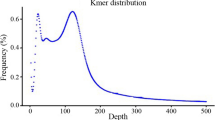
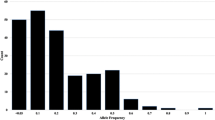
Tartary buckwheat is an important edible crop as well as medicinal plant in China. More and more research is being focused on this minor grain crop because of its medicinal functions, but there is a paucity of molecular markers for tartary buckwheat due to the lack of genomics. In this study, a genome survey was carried out in tartary buckwheat, from which SSR markers were developed for analysis of genetic diversity. The survey generated 21.9 Gb raw sequence reads which were assembled into 348.34 Mb genome sequences included 204,340 contigs. The genome size was estimated to be about 497 Mb based on K-mer analysis. In total, 24,505 SSR motifs were identified and characterised from this genomic survey sequence. Most of the SSR motifs were di-nucleotide (67.14 %) and tri-nucleotide (26.05 %) repeats. AT/AT repeat motifs were the most abundant, accounting for 78.60 % of di-nucleotide repeat motifs. SSR fingerprinting of 64 accessions yielded 49.71 effective allele loci from a total of 63 with the 23 polymorphic SSR primer combinations. Analyses of the population genetic structure using SSR data strongly suggested that the 64 accessions of tartary buckwheat clustered into two separate subgroups. One group was mainly distributed in Nepal, Bhutan and the Yunnan-Guizhou Plateau regions of China; the other group was mainly derived from the Loess Plateau regions, Hunan and Hubei of China and USA. The cluster analysis of these accession’s genetic similarity coefficient by UPMGA methods strongly supported the two subgroup interpretation. However accessions from Qinghai of China could be grouped into either of the two subgroups depending on which classification method was used. This region is at the intersection of the two geographical regions associated with the two subgroups. These results and information could be used to identify and utilize germplasm resources for improving tartary buckwheat breeding.







Similar content being viewed by others
References
Arora RK, Baniya BK, Joshi BD(1995) Buckwheat genetic resources in the Himalayas: their diversity, conservation and use. Curr Adv Buckwheat Res 39–45
Du F, Wu Y, Zhang L, Li XW, Zhao XY, Wang WH, Gao ZS, Xia YP (2015) De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium oriental hybrid ‘Sorbonne’. Plant Mol Biol Report 2:281–293
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 8:2611–2620
Fabjan N, Rode J, Kosir IJ, Wang Z, Zhang Z, Kreft I (2003) Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J Agric Food Chem 51:6452–6455
Gupta N, Sharma SK, Rana JC, Chauhan RS (2012) AFLP fingerprinting of tatary buckwheat accessions (Fagopyrum tataricum) displaying rutin content variation. Fitoterapia 83:1131–1137
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Iwata H, Imon K, Tsumura Y, Ohsawa R (2005) Genetic diversity among Japanese indigenous common buckwheat (Fagopyrum esculentum) cultivars as determined from amplified fragment length polymorphism and simple sequence repeat markers and quantitative agronomic traits. Genome 48:367–377
Kishore G, Pandey A, Dobhal R, Gupta S (2013) Population genetic study of Fagopyrum tataricum from Western Himalaya using ISSR markers. Biochem Genet 51:750–765
Lee CC, Shen SR, Lai YJ, Wu SC (2013) Rutin and quercetin, bioactive compounds from tatary buckwheat, prevent liver inflammatory injury. Food Funct 4:794–802
Logacheva MD, Penin AA, Sutormin RA, Demidenko NV, Naumenko SA, Vinogradov DV, Mazin PV, Kurmangaliev YZ, Gelfand MS, Kondrashov AS (2014) A draft genome sequence of Tatary Buckwheat, Fagopyrum tataricum. Plant & animal genome XXII, San Diego, CA. p 667
Mardis ER (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24:133–141
Nagano M, Aii J, Campbell C, Kawasaki S, Adachi T (2000) Genome size analysis of the genus Fagopyrum. Fagopyrum 17:35–39
Ohnishi O (1998) Search for the wild ancestor of buckwheat III: the wild ancestor of cultivated common buckwheat, and of Tatary buckwheat. Econ Bot 52:123–133
Parida SK, Yadava DK, Mohapatra T (2010) Microsatellites in Brassica unigenes: relative abundance, marker design, and use in comparative physical mapping and genome analysis. Genome 53:55–67
Porebski S, Bailey GL, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Report 15:8–15
Powell W, Morgante M, Andre C, Hanafey M, Vogel J (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) marker for germplasm analysis. Mol Breed 2:225–238
Sangma SC, Chrungoo NK (2010) Buckwheat gene pool: potentialities and drawbacks for use in crop improvement programmes. Eur J Plant Sci Biotechnol 4:45–50
Sharma R, Jana S (2002) Species relationships in Fagopyrum revealed by PCR-based DNA fingerprinting. Theor Appl Genet 105:306–312
Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN, Rana JC, Singh NK, Sharma TR (2011) Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One 6:e21298
Tan ML, Wu K, Wang L, Yan MF, Zhao ZD, Xu J, Zeng Y, Zhang XK, Fu CL, Xue JF, Wang LJ, Yan XC (2014) Developing and characterising Ricinus communis SSR markers by data mining of whole-genome sequences. Mol Breed 34:893–904
Tsuji K, Ohnishi O (2001) Phylogenetic relationships among wild and cultivated Tartary buckwheat (Fagopyrum tataricum Gaertn.) populations revealed by AFLP analyses. Genes Genet Syst 76:47–52
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115
Wang X, Feng B, Xu Z, Sestili F, Zhao G, Xiang C, Lafiandra D, Wang T (2014) Identification and characterization of granule bound starch synthase I (GBSSI) gene of tatary buckwheat (Fagopyrum tataricum Gaertn.). Gene 534:229–235
Yasui Y, Wang Y, Ohnishi O, Campbell CG (2004) Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self-pollinated relative Fagopyrum homotropicum. Genome 47:345–351
Zhao G, Shaan F (2008) Chinese Tartary buckwheat. Science, Beijing, pp 85–88
Zhao HS, Yang L, Peng ZH, Sun HY, Yue XH, Lou YF, Dong LL, Wang LL, Gao ZM (2015) Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci Rep 5:8018
Zietkiewicz E, Rafalski A, Labuda DF (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
We would like to thank Professor Donald Grierson, University of Nottingham, UK, for discussion and helping with the manuscript. We would like to thank Germplasm Resources Information Network in the US, Chinese Crop Germplasm Resources Information System and Institute of crop variety resources, Shanxi, China. This work was supported by NSFC (National Natural Science Foundation of China) (No. 31301385, 31471556), Science and Technology Project of Shanxi Province (20150311007-1), the Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (STIP, 2013115), the Basic Facility Platform Project of Shanxi Provincial Department of Science and Technology (2012091004–0103), Shanxi Agricultural University breeding innovation funds (2010028) and the Key Discipline Foundation of Shanxi Province.
Conflicts of Interest
The authors declare no conflict of interest.
Authors’ Contributions
Siyu Hou, Hongying Li and Yuanhuai Han designed the experiments and drafted the manuscript. Zhaoxia Sun , Lijun Zhang, Zhijun Qiao and Bin Wu collected plant materials. Bin Linghu performed the SSR experiments and analyzed the data. Dongmei Xu modified the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hou, S., Sun, Z., Linghu, B. et al. Genetic Diversity of Buckwheat Cultivars (Fagopyrum tartaricum Gaertn.) Assessed with SSR Markers Developed from Genome Survey Sequences. Plant Mol Biol Rep 34, 233–241 (2016). https://doi.org/10.1007/s11105-015-0907-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0907-5