Abstract
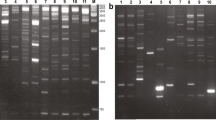
Retrotransposon-based molecular markers were applied for the first time within the genera Xanthosoma and Colocasia to assess intraspecific variability among 27 accessions of cocoyam (Xanthosoma sagittifolium) and taro (Colocasia esulenta). Retrotransposons were isolated and sequenced; long terminal repeat (LTR) primers were designed to obtain inter-retrotransposon amplified polymorphism (IRAP) fingerprints. A set of six chosen LTR primers yielded 433 reproducible bands across 20 X. sagittifolium samples. Out of the 433 bands, 400 fragments (92 %) were polymorphic. In seven C. esculenta accessions, the six primers amplified a total of 354 reproducible, informative data points, of which 285 (80.5 %) were polymorphic. Cluster analysis placed all the accessions in two groups according to their species. The accessions of X. sagittifolium were further divided into two subgroups corresponding to their ploidy level. Moreover, the genetic variability accessed by IRAP markers allowed separation of X. sagittifolium and C. esculenta accessions according to their type and botanical variety respectively. The results suggest that retrotransposon activity continued after Xanthosoma speciation. The data and approach provides a basis for better germplasm management, future systematic studies and genetic improvement, as well as for exploration of the role of retrotransposons in cocoyam and taro polyploid formation and genome dynamics.


Similar content being viewed by others
Abbreviations
- IRAP:
-
Inter-retrotransposon amplified polymorphism
- TEs:
-
Transposable elements
- LTR:
-
Long terminal repeat
- PBS:
-
Primer-binding site
- CRRD:
-
Cocoyam root rot disease
References
Adiobo A, Doungous O, Perneel M, Zok S, Höfte M (2007) Variation of Pythium-induced cocoyam root rot severity in response to soil type. Soil Biol Biochem 39:2915–2925
Alavi-Kia SS, Mohammadi SA, Aharizad S, Moghaddam M (2008) Analysis of genetic diversity and phylogenetic relationships in Crocus genus of Iran using inter-retrotransposon amplified polymorphism. Biotechnol Biotechnol Equip 22:795–800
Antonius-Kemola K, Kalendar R, Schulman AH (2006) TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor Appl Genet 112:999–1008
Baumel A, Ainouche M, Kalendar R, Schulman AH (2002) Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol Biol Evol 19:1218–1227
Benachenhou F, Sperber GO, Bongcam-Rudloff E, Andersson G, Boeke JD, Blomberg J (2013) Conserved structure and inferred evolutionary history of long terminal repeats (LTRs). Mob DNA 4:5
Boudjeko T, Omokolo ND, Driouich A, Balangé AP (2005) Peroxydase and pectin methylesterase activities in cocoyam (Xanthosoma sagittifolium L. Schott) roots upon Pythium myriotylum inoculation. J Phytopathol 153:409–416
Bown D (2000) Aroids: plants of the Arum family, 2nd edn. Timber Press, Portland
Brown VM, Asemota H (2009) PCR-based characterization of dasheen (Colocasia sp.) and cocoyam (Xanthosoma sp.). J Biotech Res 1:28–40
Cathebras C, Traore R, Malapa R, Risterucci AM, Chaïr H (2014) Characterization of microsatellites in Xanthosoma sagittifolium (Araceae) and cross-amplification in related species. Appl Plant Sci 2:1400027
Doungous O, Sama AE, Adiobo A, Zok S (2011) Determination of ploidy level by flow cytometry and autopolyploid induction in cocoyam (Xanthosoma sagittifolium). Afr J Biotechnol 10:16491–16494
FAOstat (2014) Food and Agriculture Organization of the United Nations. Rome, Italy. http://faostat3.fao.org/home/E. Accessed 30 Oct 2014
GenStat Discovery 4th edn 4 (2012)VSN International Ltd., Hemel Hempstead
Ghosh P, Mukherjee S, Sharma AK (2001) Cytophotometric estimation of in situ DNA content in several species of Araceae. Cytobios 105:177–183
Giacometti DC, Leon J (1994) Tannia, yautia (Xanthosoma sagittifolium). In: Hernando Bermejo JE, Leon J (eds) Neglected Crops: 1492 from a different perspective. FAO Plant Production and Protection Service, Rome
Grover CE, Yu Y, Wing RA, Paterson AH, Wendel JF (2008) A phylogenetic analysis of indel dynamics in the cotton genus. Mol Biol Evol 25:1415–1428
Hazzouri KM, Mohajer A, Dejak SI, Otto SP, Wright SI (2008) Contrasting patterns of transposable-element insertion polymorphism and nucleotide diversity in autotetraploid and allotetraploid Arabidopsis species. Genetics 179:581–592
Irwin SV, Kaufuis P, Banks K, de la Pena R, Cho JJ (1998) Molecular characterization of taro (Colocasia esculenta) using RAPD markers. Euphytica 99:183–199
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nature Protoc 1:2478–2484
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman AH (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA 105:5833–5838
Kalendar R, Antonius K, Smykal P, Schulman AH (2010) iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor Appl Genet 121:1419–1430
Kalendar R, Flavell A, Ellis THN, Sjakste T, Moisy C, Schulman AH (2011a) Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106:520–530
Kalendar R, Lee D, Schulman AH (2011b) Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics 98:137–144
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K (2010) Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186:801–812
Kreike CM, Van Eck HJ, Lebot V (2004) Genetic diversity of taro, Colocasia esculenta (L.) Schott, in Southeast Asia and the Pacific. Theor Appl Genet 109:761–768
Loh JP, Kiew R, Hay A, Kee A, Gan LH, Gan YY (2000) Intergeneric and interspecific relationships in Araceae tribe Caladieae and development of molecular markers using amplified fragment length polymorphism (AFLP). Ann Bot 85:371–378
Mbouobda HD, Boudjeko T, Djocgoue PF, Tsafack TJJ, Omokolo DN (2007) Morphological characterization and agronomic evaluation of cocoyam (Xanthosoma sagittifolium L. Schott) germplasm in Cameroon. J Biol Sci 7:27–33
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Nagy S, Poczai P, Cernák I, Gorji AM, Taller GHJ (2012) PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem Genet 50:670–672
Ngouo LV, Nzietchueng S, Valet G (1989) Organisation inflorentielle et florale, fertilité et germination des pollens chez trois Xanthosoma spp. cultivés au Cameroun. Agro Afr 1:95–104
Nyochembeng LM, Beyl CA, Pacumbaba RP (2007) Peroxydase activity, isozyme patterns and electrolyte leakage in roots of cocoyam infected with Pythium myriotylum. J Phytopathology 115:454–461
Offei SK, Asante IK, Danquah EY (2004) Genetic structure of seventy cocoyam (Xanthosoma sagittifolium, Linn, Schott) accessions in Ghana based on RAPD. Hereditas 140:123–128
Onokpise OU, Wutoh JG, Ndzana X, Tambong JT, Meboka MM, Sama AE, Nyochembeng L, Agueguia A, Nzietchueng S, Wilson JG, Bursn M (1999) Evaluation of macabo cocoyam germplasm in Cameroon. In: Janick J (ed) Perspectives on News Crops and New Uses. ASHA Press, Alexandra, pp 394–396
Owusu-Darko PG, Paterson A, Omenyo EL (2014) Cocoyam (corms and cormels)—An underexploited food and feed resource. J Agric Chem Environ 3:22–29
Pacumbaba RP, Wutoh JG, Sama AE, Tambong JT, Nyochembeng LM (1992) Isolation and pathogenicity of rhizosphere fungi of cocoyam in relation to cocoyam root rot disease. J Phytopathol 135:265–273
Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, Chalhoub B, Grandbastien MA (2010) Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol 186:37–45
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlex 6.5: genetic analysis in Excel. Population genetic software for teaching and research -an update. Bioinformatics 28:2537–2539
Pearce SR, Stuart-Rogers C, Knox MR, Kumar A, Ellis THN, Flavell AJ (1999) Rapid isolation of plant Ty1-copia group retrotransposon LTR sequences for molecular marker studies. Plant J 19:711–717
Perneel M, Tambong JT, Adiobo A, Floren C, Saborio F, Levesque A, Höfte M (2006) Intraspecific variability of Pythium myriotylum isolates from cocoyam and the host crops. Mycol Res 110:583–593
Perneel M, Heyrman J, Adiobo A, De Maeyer K, Raaijimakers JM, De Vos P, Höfte M (2007) Characterization of CMR5c and CMR12a, novel fluorescent Pseudomonas strains from the cocoyam rhizosphere with biocontrol activity. J Appl Microbiol 103:1007–1020
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–248
Purseglove JW (1972) Tropical crops: monocotyledons I. Longman Ltd., London
Quero-Garcia J, Noyer JL, Perrier X, Marchand JL, Lebot V (2004) A germplasm stratification of taro (Colocasia esculenta) based on agro-morphological descriptors, validation by AFLP markers. Euphytica 137:387–395
Schlüter PM, Harris SA (2006) Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes 6:569–572
Schnell RJ, Goenaga R, Olano CT (1999) Genetic similarity among cocoyam cultivars based on randomly amplified polymorphic DNA (RAPD) analysis. Sci Hortic 80:267–276
Schulman AH (2013) Retrotransposon replication in plants. Curr Opin Virol 3:604–614
Schulman A, Flavell A, Paux E, Ellis THN (2012) The application of LTR retrotransposons as molecular markers in plants. Methods Mol Biol 859:115–153
Sefa-Dedeh S, Agyir-Sackey KE (2004) Chemical composition and effect of processing on oxalate content of cocoyam Xanthosoma sagittifolium and Colocasia esculenta cormels. Food Chem 85:479–487
Shannon CE, Weaver W (1949) The mathematical theory of communication. The University of Illinois Press, Urbana
Singh D, Jackson G, Hunter D, Fullerton R, Lebot V, Taylor M, Iosefa T, Okpul T, Tyson J (2012) Taro leaf blight—a threat to food security. Agriculture 2:182–203
Smykal P, Bacova-kerteszova N, Kalendar R, Corander J, Schulman AH, Pavelek M (2011) Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor Appl Genet 122:1385–1397
Swofford DL (1998) PAUP. Phylogenetic analysis using parsimony (and other methods). Version 4. Sinauer Associates, Sunderland
Tambong JT, Sapra VT, Garton S (1998) In vitro induction of tetraploids in colchicine-treated cocoyam plantlets. Euphytica 104:191–197
Vicient CMM, Jääskeläinen M, Kalendar R, Schulman AH (2001) Active retrotransposons are a common feature of grass genomes. Plant Physiol 125:1283–1292
Vukich M, Schulman AH, Giordani T, Natali L, Kalendar R, Cavallini A (2009) Genetic diversity in sunflower (Helianthus annus L.) and the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet 119:1027–1038
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wilson JE (1984) Cocoyam. In: Goldsworthy PR, Fisher NM (eds) The physiology of tropical field crops. Wiley, London, pp 589–605
Yeh FC, Boyle TBJ (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129:157
Acknowledgments
Anne-Mari Narvanto and Ursula Lönnqvist are thanked for their valuable technical assistance. D.O. was supported by an 8 month-mobility fellowship (Decision TM-09-6259) and a travel grant issued by the Finnish Centre for International Mobility (CIMO) and the Kirkhouse Trust respectively. R.K. was supported by a grant from the Academy of Finland (Decision 134079).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest. The experiments comply with the current laws of the countries in which they were performed.
Additional information
Data deposition The sequences reported here have been deposited in the GenBank database (accession nos.GU810535, KF813039–KF813057).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Doungous, O., Kalendar, R., Adiobo, A. et al. Retrotransposon molecular markers resolve cocoyam (Xanthosoma sagittifolium) and taro (Colocasia esculenta) by type and variety. Euphytica 206, 541–554 (2015). https://doi.org/10.1007/s10681-015-1537-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1537-6