Abstract
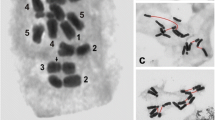
Carrot (Daucus carota L.) chromosomes are small and uniform in shape and length. Here, mitotic chromosomes were subjected to multicolour fluorescence in situ hybridization (mFISH) with probes derived from conserved plant repetitive DNA (18-25S and 5S rDNA, telomeres), a carrot-specific centromeric repeat (Cent-Dc), carrot-specific repetitive elements (DCREs), and miniature inverted-repeat transposable elements (MITEs). A set of major chromosomal landmarks comprising rDNA and telomeric and centromeric sequences in combination with chromosomal measurements enabled discrimination of carrot chromosomes. In addition, reproducible and unique FISH patterns generated by three carrot genome-specific repeats (DCRE22, DCRE16, and DCRE9) and two transposon families (DcSto and Krak) in combination with telomeric and centromeric reference probes allowed identification of chromosome pairs and construction of detailed carrot karyotypes. Hybridization patterns for DCREs were observed as pericentromeric and interstitial dotted tracks (DCRE22), signals in pericentromeric regions (DCRE16), or scattered signals (DCRE9) along chromosomes similar to those observed for both MITE families.
Similar content being viewed by others
Abbreviations
- BAC:
-
bacterial artificial chromosome
- DAPI:
-
4′,6-diamidino-2-phenylindole
- DCRE:
-
Daucus carota repetitive element
- FISH:
-
fluorescence in situ hybridization
- FITC:
-
fluorescein isothiocyanate
- mFISH:
-
multicolour fluorescence in situ hybridization
- MITE:
-
miniature inverted-repeat transposable element
- NOR:
-
nucleolar organizer region
- SSC:
-
saline-sodium citrate
- TE:
-
transposable element
- TIR:
-
terminal inverted repeat
- TRS:
-
telomeric repeat sequence
- TSD:
-
target site duplications
References
Altinkut, A., Kotseruba, V., Kirzhner, V.M., Nevo, E., Raskina, O., Belyayev, A.: Ac-like transposons in populations of wild diploid Triticeae species: comparative analysis of chromosomal distribution. — Chromosome Res. 14: 307–317, 2006.
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped BLAST and PSIBLAST; a new generation of protein database search programs. — Nucl. Acids Res. 25: 3389–3402, 1997.
Arscott, S.A., Tanumihardjo, S.A.: Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. — Comp. Rev. Food Sci. Safety 9: 223–239, 2010.
Arumuganathan, K., Earle, E.D.: Nuclear DNA content of some important plant species. — Plant mol. biol. Rep. 9: 208–218, 1991.
Boyle, S., Rodesch, M.J., Halvensleben, H.A., Jeddeloh, J.A., Bickmore, W.A.: Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. — Chromosome Res. 19: 901–909, 2011.
Cavagnaro, P.F., Chung, S.M., Szklarczyk, M., Grzebelus, D., Senalik, D., Atkins, A.E., Simon, P.W.: Characterization of a deep-coverage carrot (Daucus carota L.) BAC library and initial analysis of BAC-end sequences. — Mol. Genet. Genomics 281: 273–288, 2009.
Cheng, Z., Dong, F., Langdon, T., Ouyang, S., Buell, R., Gu, M., Blattner, F.R., Jiang, J.: Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. — Plant Cell 14: 1691–1704, 2002.
Chomczynski, P., Sacchi, N.: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenolchloroform extraction. — Anal. Biochem. 162: 156–159, 1987.
Dechyeva, D., Gindullis, F., Schmidt, T.: Divergence of satellite DNA and interspersion of dispersed repeats in the genome of the wild beet Beta procumbens. — Chromosome Res. 11; 3–21, 2003.
Dydak, M., Kolano, B., Nowak, T., Siwinska, D,. Maluszynska, J.: Cytogenetic studies of three European species of Centaurea L. (Asteraceae). — Hereditas 146: 152–161, 2009.
Essad, S.: [Banding and biometry applied to karyotype analysis in Daucus carota L.] — Agronomie 5: 871–876, 1985. [In French]
Feschotte, C., Swamy, L., Wessler, S.R.: Genome-wide analysis of Mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). — Genetics 163: 747–758, 2003.
Findley, S.D., Cannon, S., Varala, K., Du, J., Ma, J., Hudson, M.E., Birchler, J.A., Stacey, G.: A fluorescence in situ hybridization system for karyotyping soybean. — Genetics 185: 727–744, 2010.
Fuchs, J., Brandes, A., Schubert, I.: Telomere sequence localization and karyotype evolution in higher plants. — Plant Syst. Evol. 196: 227–241, 1995.
Gerlach, W.L., Bedbrook, J.R.: Cloning and characterization of ribosomal RNA genes from wheat and barley. — Nucl. Acids Res. 109: 1346–1352, 1979.
Gerlach, W.L., Dyer, T.A.: Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. — Nucl. Acids Res. 8: 4851–4865, 1980.
Grabowska-Joachimiak, A., Kula, A., Gernand-Kliefoth, D., Joachimiak, A.J.: Karyotype structure and chromosome fragility in the grass Phleum echinatum Host. — Protoplasma 252: 301–306, 2015.
Grzebelus, D., Jagosz, B., Simon, P.W.: The DcMaster transposon display maps polymorphic insertion sites in the carrot (Daucus carota L.) genome. — Gene 390: 67–74, 2007.
Grzebelus, D., Simon, P.W.: Diversity of DcMaster-like elements of the PIF/Harbinger superfamily in the carrot genome. — Genetica 135: 347–353, 2009.
Grzebelus, D., Yau, Y.-Y., Simon, P.W.: Master: a novel family of PIF/Harbinger-like transposable elements identified in carrot (Daucus carota L.). Mol. Genet. Genomics 275: 450–459, 2006.
Hajdera, I., Siwinska, D,. Hasterok, R., Maluszynska, J.; Molecular cytogenetic analysis of genome structure in Lupinus angustifolius and Lupinus cosentinii. — Theor. appl. Genet. 107: 988–996, 2003.
Hasterok, R., Jenkins, G., Langdon, T., Jones, R.N., Maluszynska, J.: Ribosomal DNA is an effective marker of Brassica chromosomes. — Theor. appl. Genet. 103: 486–490, 2001.
Hasterok, R., Langdon, T., Taylor, S., Jenkins, G.; Combinatorial labelling of DNA probes enables multicolour fluorescence in situ hybridisation in plants. — Folia histochem. cytobiol. 40: 319–332, 2002.
Heslop-Harrison, J.S.: Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. — Plant Cell 12: 617–635, 2000.
Heslop-Harrison, J.S., Brandes, A., Taketa, S., Schmidt, T., Vershinin, A.V., Alkhimova, E.G., Kamm, A., Doudrick, R.L., Schwarzacher, T., Katsiotis, A., Kubis, S., Kumar, A., Pearce, S.R., Flavell, A.J., Harrison, G.E.: The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. — Genetica 100: 197–204, 1997.
Hizume, M., Shibata, F., Matsusaki, Y., Garajova, Z.; Chromosome identification and comparative karyotypic analyses of four Pinus species. — Theor. appl. Genet. 105; 491–497, 2002.
Horakova, M., Fajkus, J.: TAS49 - a dispersed repetitive sequence isolated from subtelomeric regions of Nicotiana tomentosiformis chromosomes. — Genome 43: 273–284, 2000.
Hoshi, Y., Yagi, K., Matsuda, M., Matoba, H., Tagashira, N., Plader, W., Malepszy, S., Nagano, K., Morikawa, A.: A comparative study of the three cucumber cultivars using fluorescent staining and fluorescence in situ hybridization. — Cytologia 76: 3–10, 2011.
Hueros, G., Loarce, Y., Ferrer, E.: A structural and evolutionary analysis of a dispersed repetitive sequence. — Plant mol. Biol. 22: 635–643, 1993.
Idziak, D., Hazuka, I., Poliwczak, B., Wiszynska, A., Wolny, E., Hasterok, R.: Insight into the karyotype evolution of Brachypodium species using comparative chromosome barcoding. — PLoS ONE 9: e93503, 2014.
Iovene, M., Grzebelus, E., Carputo, D., Jiang, J., Simon, P.W.; Major cytogenetic landmarks and karyotype analysis in Daucus carota and other Apiaceae. — Amer. J. Bot. 95: 793–804, 2008.
Iovene, M., Cavagnaro, P.F., Senalik, D., Buell, C.R., Jiang, J., Simon, P.W.: Comparative FISH mapping of Daucus species (Apiaceae family). — Chromosome Res. 19: 493–506, 2011.
Itoh, Y., Hasebe, M., Davies, E., Takeda, J., Ozeki, Y.: Survival of Tdc transposable elements of the En/Spm superfamily in the carrot genome. — Mol. Genet. Genomics 269: 49–59, 2003.
Jiang, N., Feschotte, C., Zhang, X., Wessler, S.R.: Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). — Curr. Opin. Plant Biol. 7: 115–119, 2004.
Jin, W., Melob, J.R., Nagaki, K., Talbert, P.B., Henikoff, S., Kelly, D.R., Jiang, J.: Maize centromeres: organization and functional adaptation in the genetic background of oat. —Plant Cell 16: 571–581, 2004.
Jurka, J., Kapitonov, V.V., Pavlicek, A., Klonowski, P., Kohany, O., Walichiewicz, J.: Repbase update, a database of eukaryotic repetitive elements. — Cytogenet. Genome Res. 110: 462–467, 2005.
Kato, A., Lamb, J.C., Birchler, J.A.: Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. — Proc. nat. Acad. Sci. USA 101: 13554–13559, 2004.
Kiefer-Meyer, M.C., Reddy, A.S., Delseny, M.: Complex arrangement of dispersed repeated DNA sequences in Oryza officinalis. — Genome 39: 183–190, 1996.
Kolano, B., Plucienniczak, A., Kwasniewski, M., Maluszynska, J.: Chromosomal localization of a novel repetitive sequence in the Chenopodium quinoa genome. — J. appl. Genet. 49; 313–320, 2008.
Kolano, B., Gardunia, B.W., Michalska, M., Bonifacio, A., Fairbanks, D., Maughan, P.J., Coleman, C.E., Stevens, M.R., Jellen, E.N., Maluszynska, J.: Chromosomal localization of two novel repetitive sequences isolated from the Chenopodium quinoa Willd. — Genome 54: 710–717, 2011.
Kubis, S., Schmidt, T., Seymour, J., Heslop-Harrison, J.S.; Repetitive DNA elements as a major component of plant genomes. — Ann. Bot. 82: 45–55, 1998.
Kulikova, O., Gualtieri, G., Guerts, R., Kim, D.J., Cook, D.; Integration of the FISH pachytene and genetic maps of Medicago truncatula. — Plant J. 27: 49–58, 2001.
Kulikova, O., Geurts, R., Lamine, M., Kim, D.J., Cook, D.R., Leunissen, J., De Jong, H., Roe, B.A., Bisseling, T.; Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicago truncatula. — Chromosoma 113: 276–283, 2004.
Kumar, P., Widholm, J.M.: Techniques for chromosome analysis of carrot culture cells. — Plant mol. biol. Rep. 2: 37–42, 1984.
Kumar, A., Bennetzen, J.L.: Plant retrotransposons. — Annu. Rev. Genet. 33: 479–532, 1999.
Levan, A., Fredga, K., Sandberg, A.A.: Nomenclature for centromeric position on chromosomes. — Hereditas 52: 201–220, 1964.
Macko-Podgorni, A., Nowicka, A., Grzebelus, E., Simon, P.W., Grzebelus, D.: DcSto: carrot Stowaway-like elements are abundant, diverse, and polymorphic. — Genetica 141: 255–267, 2013.
Menzel, G., Dechyeva, D., Keller, H., Lange, C., Himmelbauer, H., Schmidt, T.: Mobilization and evolutionary history of miniature inverted-repeat transposable elements (MITEs) in Beta vulgaris L. — Chromosome Res. 14: 831–844, 2006.
Menzel, G., Dechyeva, D., Wenke, T., Holtgraewe, D., Weisshaar, B., Schmidt, T.: Diversity of a complex centromeric satellite and molecular characterization of dispersed sequence families in sugar beet (Beta vulgaris). — Ann. Bot. 102: 521–530, 2008.
Navratilova, A., Neumann, P., Macas, J.: Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. — Ann. Bot. 91: 921–926, 2003.
Neumann, P., Nouzova, M., Macas, J.: Molecular and cytogenetic analysis of repetitive DNA in pea (Pisum sativum L.). — Genome 44: 716–728, 2001.
Nowicka, A., Grzebelus, E., Grzebelus, D.: Fluorescent in situ hybridization with arbitrarily amplified DNA fragments differentiates carrot (Daucus carota L.) chromosomes. —Genome 55: 205–213, 2012.
Ozeki, Y., Davies, E., Takeda, J.: Somatic variation during long term subculturing of plant cells caused by insertion of a transposable element in a phenylalanine ammonia-lyase (PAL) gene. — Mol. gen. Genet. 254: 407–416, 1997.
Paesold, S., Borchardt, D., Schmidt, T., Dechyeva, D.: A sugar beet (Beta vulgaris L.) reference FISH karyotype for chromosome and chromosome-arm identification, integration of genetic linkage groups and analysis of major repeat family distribution. — Plant J. 72: 600–611, 2012.
Rozen, S., Skaletsky, H.J.: Primer3 on the www for general users and for biologist programmers. — In: Krawetz, S., Misener, S. (ed.): Bioinformatics Methods and Protocols; Methods in Molecular Biology. Pp. 365–386. Humana Press, Totowa 2000.
Schmidt, T., Kubis, S., Katsiotis, A., Jung, C., Heslop-Harrison, J.S.: Molecular and chromosomal organization of two repetitive DNA sequences with intercalary locations in sugar beet and other Beta species. — Theor. appl. Genet. 97; 696–704, 1998.
Schrader, O., Ahne, R., Fuchs, J.: Karyotype analysis of Daucus carota L. using Giemsa C-Banding and FISH of 5S and 18S/25S rRNA specific genes. — Caryologia 56: 149–154, 2003.
Schwarzacher, T.: DNA, chromosomes, and in situ hybridization. — Genome 46: 953–962, 2003.
Stebbins, G.L.: Chromosomal Evolution in Higher Plants. — Edward Arnold Publishers, London 1971.
Szinay, D., Chang, S.B., Khrustaleva, L., Peters, S., Schijlen, E., Bai, Y., Stiekema, W.J., Van Ham, R., De Jong, H., Klein Lankhorst, R.: High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. — Plant J. 56: 627–637, 2008.
Turcotte, K., Srinivasan, S., Bureau, T.E.: Survey of transposable elements from rice genomic sequences. — Plant J. 25: 169–179, 2001.
Weber, B., Wenke, T., Frommel, U., Schmidt, T., Heitkam, T.; The Ty1-copia families SALIRE and Cotzilla populating the Beta vulgaris genome show remarkable differences in abudance, chromosomal distribution, and age. — Chromosome Res. 18: 247–263, 2010.
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J.L., Capy, P., Chalhoub, B., Flavell, A., Leroy, P., Morgante, M., Panaud, O., Paux, E., San Miguel, P., Schulman, A.H.: A unified classification system for eukaryotic transposable elements. — Nat. Rev. Genet. 8: 973–982, 2007.
Wolny, E., Fidyk, W., Hasterok, R.: Karyotyping of Brachypodium pinnatum (2n = 18) chromosomes using cross-species BAC-FISH. — Genome 56: 239–243, 2013.
Yu, W., Lamb, J.C., Han, F., Birchler, J.A.: Cytological visualization of DNA transposons and their transposition pattern in somatic cells of maize. — Genetics 175: 31–39, 2007.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgments: This research was financed by the Ministry of Science and Higher Education of the Republic of Poland.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nowicka, A., Grzebelus, E. & Grzebelus, D. Precise karyotyping of carrot mitotic chromosomes using multicolour-FISH with repetitive DNA. Biol Plant 60, 25–36 (2016). https://doi.org/10.1007/s10535-015-0558-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-015-0558-2