Abstract
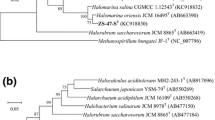
A halophilic archaeal strain, designated ZS-57-ST, was isolated from Zhoushan marine solar saltern, China. Cells were observed to be pleomorphic, stained Gram-negative and formed red pigmented colonies on agar plates. Optimal growth was obtained at 3.9 M NaCl (range 1.4–4.8 M), 0.3 M MgCl2 (range 0–1.0 M), 30 °C (range 20–55 °C) and pH 6.5–7.5 (range 5.5–9.0). The cells were found to lyse in distilled water and the minimal NaCl concentration to prevent cell lysis was determined to be 5 % (w/v). The major polar lipids were identified as C20C20 and C20C25 diether derivatives of phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, phosphatidylglycerol sulfate, glucosyl mannosyl glucosyl diether and two unidentified glycolipids. The 16S rRNA gene and rpoB′ gene of strain ZS-57-ST were phylogenetically related to the corresponding genes of Halomarina oriensis JCM 16495T (98.2 and 93.7 % similarities, respectively). The DNA G+C content of strain ZS-57-ST was determined to be 67.1 mol% (T m). The phenotypic, chemotaxonomic and phylogenetic properties suggested that strain ZS-57-ST (=CGMCC 1.12543T = JCM 30039T) represents a new species of the genus Halomarina, for which the name Halomarina salina sp. nov. is proposed.

Similar content being viewed by others
References
Cui H-L, Lin Z-Y, Dong Y, Zhou P-J, Liu S-J (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206
Cui H-L, Zhou P-J, Oren A, Liu S-J (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13:31–37
Cui H-L, Gao X, Yang X, Xu X-W (2010) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14:493–499
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Gupta RS, Naushad S, Baker S (2015) Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol 65:1050–1069
Gutiérrez C, González C (1972) Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl Microbiol 24:516–517
Henriet O, Fourmentin J, Delincé B, Mahillon J (2014) Exploring the diversity of extremely halophilic archaea in food-grade salts. Int J Food Microbiol 191:36–44
Inoue K, Itoh T, Ohkuma M, Kogure K (2011) Halomarina oriensis gen. nov., sp. nov., a halophilic archaeon isolated from a seawater aquarium. Int J Syst Evol Microbiol 61:942–946
Ivanov VM (2004) The 125th anniversary of the Griess reagent. J Anal Chem 59:1002–1005
Kim OS, Cho YJ, Lee K, Yoon S-H, Kim M, Na H, Park S-C, Jeon YS, Lee J-H, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim M, Oh H-S, Park S-C, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Liu Q, Ren M, Zhang L-L (2015) Natribaculum breve gen. nov., sp. nov. and Natribaculum longum sp. nov., halophilic archaea isolated from saline soil. Int J Syst Evol Microbiol 65:604–608
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
McDade JJ, Weaver RH (1959) Rapid methods for the detection of gelatin hydrolysis. J Bacteriol 77:60–64
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B’ (rpoB′) gene. Int J Syst Evol Microbiol 60:2398–2408
Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE (2012) De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6:81–93
Oren A (2014a) Taxonomy of halophilic Archaea: current status and future challenges. Extremophiles 18:825–834
Oren A (2014b) Family Halobacteriaceae. In: Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E (eds) The prokaryotes. A handbook on the biology of bacteria: ecophysiology and biochemistry, 4th edn. Springer, New York
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Purdy KJ, Cresswell-Maynard TD, Nedwell DB, McGenity TJ, Grant WD, Timmis KN, Embley TM (2004) Isolation of haloarchaea that grow at low salinities. Environ Microbiol 6:591–595
Rodriguez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A (1979) Isolation of extreme halophiles from seawater. Appl Environ Microbiol 38:164–165
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Savage KN, Krumholz LR, Oren A, Elshahed MS (2007) Haladaptatus paucihalophilus gen. nov., sp. nov., a halophilic archaeon isolated from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol 57:19–24
Sorokin DY, Toshchakov SV, Kolganova TV, Kublanov IV (2015) Halo(natrono)archaea isolated from hypersaline lakes utilize cellulose and chitin as growth substrates. Front Microbiol 6:942
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 30:2725–2729
Viver T, Cifuentes A, Díaz S, Rodríguez-Valdecantos G, González B, Antón J, Rosselló-Móra R (2015) Diversity of extremely halophilic cultivable prokaryotes in Mediterranean, Atlantic and Pacific solar salterns: Evidence that unexplored sites constitute sources of cultivable novelty. Syst Appl Microbiol 38:266–275
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31370054), the opening project of State key Laboratory of Microbial Resources (No. SKLMR-20150603, Institute of Microbiology, Chinese Academy of Sciences) and the 11th “Six Talents Peak” Project of Jiangsu Province (No. 2014-SWYY-021).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, WM., Xu, JQ., Zhou, Y. et al. Halomarina salina sp. nov., isolated from a marine solar saltern. Antonie van Leeuwenhoek 109, 1121–1126 (2016). https://doi.org/10.1007/s10482-016-0714-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0714-5