Abstract
Chinese Luzhou-flavor liquor is prepared using distillation of mixed, fermented grains, such as sorghum and rice. Fermentation occurs in cellars lined with pit mud containing microbes. The identity and community diversity of bacteria in pit mud of fermentation cellars contributes to the aromatic compounds of the liquor. In order to determine the contribution of genus Clostridium to this fermentation, the community diversity of Clostridium cluster I in pit mud of different cellar ages (1, 50, 100, and 400 years) was investigated based on denaturing gradient gel electrophoresis followed by 16S rRNA sequencing. Differences in the community structure within pit mud depended on wall and bottom layer cellar locations. A better understanding of the role of Clostridium in Luzhou-flavor liquor manufacture will be useful.
Similar content being viewed by others
References
Xu Y, Wang D, Fan WL, Mu XQ, Chen J. Traditional Chinese biotechnology. Vol. 122, pp. 189–233. In: Biotechnology in China II. Tsao GT, Ouyang P, Chen J (eds). Springer-Verlag, Berlin Heidelberg, New York, NY, USA (2010)
Ding XS, Zhao H. Research progress in Clostridium kluyveri. Sci. Tech. Food Ind. 33: 401–405 (2012)
Zhao H, Chang Y, Wang W, Ling HZ, Ping WX, Zhao ZC, Yang ZH. Isolation and identification of facultative anaerobic strains with high yield of hexanoic acid from Luzhou-flavor liquor pit mud. Food Sci. 33: 177–182 (2012)
Wang LP, Zhang L, Liu LY, Zhang SY, Xu DF, Liu GY. Comparative study of two Methanobacterium strains in the ancient fermentation pits of Luzhou. Chin. J. Appl. Environ. Biol. 2010: 840–844 (2010)
Deng B, Shen CH, Shan XH, Ao ZH, Zhao JS, Shen XJ, Huang ZG. PCR-DGGE analysis on microbial communities in pit mud of cellars used for different periods of time. J. I. Brewing 118: 120–126 (2012)
Liu MK, Tang YM, Zou L, Ren DQ, Yao WC, Tian XH, Yi B, Deng B. Archaea community and diversity in the pit mud of Chinese Luzhou-flavor liquor. Food Ferment. Ind. 39: 22–28 (2013)
Wang MY, Zhang WX, Wang HY, Liu CL. Analysis of bacterial phylogenetic diversity of the pit mud with different cellar age. Food Sci. 34: 177–181 (2013)
Zheng J, Liang R, Zhang LQ, Wu CD, Zhou RQ, Liao XP. Characterization of microbial communities in strong aromatic liquor fermentation pit muds of different ages assessed by combined DGGE and PLFA analyses. Food Res. Int. 54: 660–666 (2013)
Tao Y, Li JB, Rui JP, Xu ZC, Zhou Y, Hu XH, Wang X, Liu MH, Li DP, Li XZ. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl. Environ. Microb. 80: 2254–2260 (2014)
Ding XF, Wu CD, Zhang LQ, Zheng J, Zhou RQ. Characterization of eubacterial and archaeal community diversity in the pit mud of Chinese Luzhou-flavor liquor by nested PCR-DGGE. World J. Microb. Biot. 30: 605–612 (2014)
Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. Clostridia: The importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotech. 23: 364–381 (2012)
Wang CL, Shi DJ, Gong GL. Microorganisms in Daqu: A starter culture of Chinese Maotai-flavor liquor. World J. Microb. Biot. 24: 2183–2190 (2008)
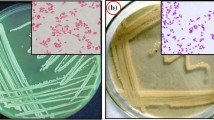
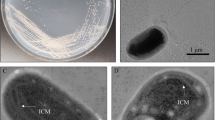
Liu CL, Huang D, Liu LY, Zhang J, Deng Y, Chen L, Zhang WX, Wu ZY, Fan A, Lai DY, Dai LR. Clostridium swellfunianum sp. nov., a novel anaerobic bacterium isolated from the pit mud of Chinese Luzhou-flavor liquor production. A. Van Leeuw. J. Microb. 106: 817–825 (2014)
Xie SG, Dai Q, Zhao SM, Liang YX. Study on isolation of Clostridium butyricum from pits sludge and its characteristics. Microbiol. China 34: 1047–1051 (2007)
Yue YY, Zhang WX, Liu X, Hu C, Zhang SY. Isolation and identification of facultative anaerobes in the pit mud of Chinese Luzhou-flavor liquor. Microbiol. China 34: 251–255 (2007)
Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int. J. Syst. Evol. Micr. 44: 812–826 (1994)
Hung CH, Cheng CH, Cheng LH, Liang CM, Lin CY. Application of Clostridium-specific PCR primers on the analysis of dark fermentation hydrogen-producing bacterial community. Int. J. Hydrogen Energ. 33: 1586–1592 (2008)
Teachcn. Top ten brands of wine in China. Available from: http:// www.teachcn.com/Content.asp?id=733. Accessed Sep. 19, 2011.
Bassam BJ, Caetano-Anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196: 80–83 (1991)
Sokal RR. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38: 1409–1438 (1958)
Dice LR. Measures of the amount of ecologic association between species. Ecology 26: 297–302 (1945)
Hill TCJ, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43: 1–11 (2003)
Li XL, Penttinen P, Gu YF, Zhang XP. Diversity of nifH gene in rhizosphere and non-rhizosphere soil of tobacco in Panzhihua, China. Ann. Microbiol. 62: 995–1001 (2012)
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 (1997)
Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 (1987)
McCann KS. The diversity-stability debate. Nature 405: 228–233 (2000)
Ren DQ, Liu MK, Tang YM, Liu XY, Yao WC, Ren JB, Tian XH, Zhang XY. Analysis of physicochemical characteristic in pit mud of Luzhou-flavor liquor. China Brewing 33: 55–57 (2014)
Ye NF, Lü F, Shao LM, Godon JJ, He PJ. Bacterial community dynamics and product distribution during pH-adjusted fermentation of vegetable wastes. J. Appl. Microbiol. 103: 1055–1065 (2007)
Kreitz S, Anderson TH. Substrate utilization patterns of extractable and non-extractable bacterial fractions in neutral and acidic beech forest soils. pp. 149–160. In: Microbial Communities. Heribert I, Andrea R (eds). Springer-Verlag, Berlin Heidelberg, New York, NY, USA (1997)
Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10: 2966–2978 (2008)
Kenealy WR, Cao Y, Weimer PJ. Production of caproic acid by cocultures of ruminal cellulolytic bacteria and Clostridium kluyveri grown on cellulose and ethanol. Appl. Microbiol. Biot. 44: 507–513 (1995)
Ren DQ, Tang YM, Yi B, Yao WC, Lu ZM, Liao JM. Promotion of quality liquor rate using microorganism technology of upper layer formation grains. China Brewing 30: 163–166 (2011)
Zhao K, Penttinen P, Chen Q, Guan TW, Lindström K, Ao XL, Zhang LL, Zhang XP. The Rhizospheres of traditional medicinal plants in Panxi, China, host a diverse selection of actinobacteria with antimicrobial properties. Appl. Microbiol. Biot. 94: 1321–1335 (2012)
Barker HA, Kamen MD, Bornstein BT. The synthesis of butyric and caproic acids from ethanol and acetic acid by Clostridium kluyveri. P. Natl. Acad. Sci. USA 31: 373–381 (1945)
Jeon BS, Kim BC, Um Y, Sang BI. Production of hexanoic acid from D-galactitol by a newly isolated Clostridium sp. BS-1. Appl. Microbiol. Biot. 88: 1161–1167 (2010)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, M., Zhao, K., Tang, Y. et al. Analysis of Clostridium cluster I community diversity in pit mud used in manufacture of Chinese Luzhou-flavor liquor. Food Sci Biotechnol 24, 995–1000 (2015). https://doi.org/10.1007/s10068-015-0127-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-015-0127-7