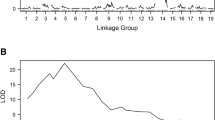
Abstract
Grapevines (Vitis vinifera L.) form the basis of viticulture, and are susceptible to diseases such as downy mildew (Plasmopara viticola) and powdery mildew (Erysiphe necator). Therefore, successful viticulture programs require the use of pesticides. Breeding for resistance is the only eco-friendly solution. Marker-assisted selection is currently widely used for grapevine breeding. Consequently, traits of interest must be tagged with molecular markers linked to quantitative trait loci (QTL). We herein present our findings regarding genetic mapping and QTL analysis of resistance to downy and powdery mildew diseases in the progenies of the GF.GA-47-42 (‘Bacchus’ × ‘Seyval’) × ‘Villard blanc’ cross. Simple sequence repeats and single nucleotide polymorphisms of 151 individuals were analyzed. A map consisting of 543 loci was screened for QTL analyses based on phenotypic variations observed in plants grown in the field or under controlled conditions. A major QTL for downy mildew resistance was detected on chromosome 18. For powdery mildew resistance, a QTL was identified on chromosome 15. This QTL was replaced by a novel QTL on chromosome 18 in 2003 (abnormally high temperatures) and 2004. Subsequently, both QTLs functioned together. Additionally, variations in the timing of the onset of veraison, which is a crucial step during grape ripening, were studied to identify genomic regions affecting this trait. A major QTL was detected on linkage group 16, which was supplemented by a minor QTL on linkage group 18. This study provides useful information regarding novel QTL-linked markers relevant for the breeding of disease-resistant grapevines adapted to current climatic conditions.




Similar content being viewed by others
References
Akkurt M, Welter L, Maul E, Töpfer R, Zyprian E (2007) Development of SCAR markers linked to powdery mildew (Uncinula necator) resistance in grapevine (Vitis vinifera L. and Vitis sp.). Mol Breeding 19:103–111
Austin CN, Wilcox WF (2012) Effects of sunlight exposure on grapevine powdery mildew development. Phytopathology 102:857–866
Barnavon L, Doco T, Terrier N, Ageorges A, Romieu C, Pellerin P (2000) Analysis of cell wall neutral sugar composition, beta-galactosidase activity and a related cDNA clone throughout the development of Vitis vinifera grape berries. Plant Physiol Biochem 38:289–300
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon AF, Cipriani G, Morgante M, Testolin R, Di Gaspero G (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet 120:163–176
Cabezas JA, Ibanez J, Lijavetzky D, Velez D, Bravo G, Rodriguez V, Carreno I, Jermakow AM, Carreno J, Ruiz-Garcia L, Thomas MR, Martinez-Zapater JM (2011) A 48 SNP set for grapevine cultivar identification. BMC Plant Biol 11:153
Choudhury RA, McRoberts N, Gubler WD (2014) Effects of punctuated heat stress on the grapevine powdery mildew pathogen, Erysiphe necator. Phytopathol Mediterr 53:148–158
Cipriani G, Marrazzo M-T, Di Gaspero G, Pfeiffer A, Morgante M, Testolin R (2008) A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol 8:127
Costantini L, Battilana J, Lamaj F, Fanizza G, Grando MS (2008) Berry and phenology-related traits in grapevine (Vitis vinifera L.): From Quantitative Trait Loci to underlying genes. BMC Plant Biol 8:38
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Deglene-Benbrahim L, Wiedemann-Merdinoglu S, Merdinoglu D, Walter B (2010) Evaluation of downy mildew resistance in grapevine by leaf disc bioassay with in vitro- and greenhouse-grown plants. Am J Enol Vitic 61:521–528
Delmotte F, Mestre P, Schneider C, Kassemeyer HH, Kozma P, Richart-Cervera S, Rouxel M, Deliere L (2014) Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect Genet Evol 27:500–508
Di Gaspero G, Copetti D, Coleman C, Castellarin SD, Eibach R, Kozma P, Lacombe T, Gambetta G, Zvyagin A, Cindric P, Kovacs L, Morgante M, Testolin R (2012) Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor Appl Genet 124:277–286
Duchene E, Butterlin G, Dumas V, Merdinoglu D (2012) Towards the adaptation of grapevine varieties to climate change: QTLs and candidate genes for developmental stages. Theor Appl Genet 124:623–635
Dudenhöffer J (2012) Diploma thesis: Physikalische Kartierung der Resistenzregion gegen den Echten Mehltau aus der Rebsorte ‘Regent’. In: Faculty of Chemistry and Life Sciences. Karlsruhe Institute of Technology (KIT), Karlsruhe
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM, Grando MS (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13:39
Fechter I, Hausmann L, Daum M, Rosleff Sörensen T, Viehöver P, Weisshaar B, Töpfer R (2012) Candidate genes within a 143 kb region of the flower sex locus in Vitis. Mol Genet Genomics 287:247–259
Fechter I, Hausmann L, Zyprian E, Daum M, Holtgräwe D, Weisshaar B, Töpfer R (2014) QTL analysis of flowering time and ripening traits suggests an impact of a genomic region on linkage group 1 in Vitis. Theor Appl Genet 127:1857–1872
Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Töpfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108:501–515
Gadoury DM, Cadle-Davidson L, Wilcox WF, Dry IB, Seem RC, Milgroom MG (2012) Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol Plant Pathol 13:1–16
Genet JL, Jaworska G (2013) Characterization of European Plasmopara viticola isolates with reduced sensitivity to cymoxanil. Eur J Plant Pathol 135:383–393
Gentry M, Hennig L (2014) Remodelling chromatin to shape development of plants. Exp Cell Res 321:40–46
Gessler C, Pertot I, Perazzolli M (2011) Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol Mediterr 50:3–44
Grattapaglia D, Sederoff R (1994) Genetic-linkage maps of Eucalyptus-grandis and Eucalyptus-urophylla using a pseudo-testcross—mapping strategy and rapd markers. Genetics 137:1121–1137
Hunt HV, Lawes MC, Bower MA, Haeger JW, Howe CJ (2010) A banned variety was the mother of several major wine grapes. Biol Lett 6:367–369
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P, The French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–465
Jeong HJ, Kim YJ, Kim SH, Kim YH, Lee IJ, Kim YK, Shin JS (2011) Nonsense-mediated mRNA Decay Factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol 52:2147–2156
Jiang RHY, Tyler BM (2012) Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol 50(50):295–318
Mahanil S, Ramming D, Cadle-Davidson M, Owens C, Garris A, Myles S, Cadle-Davidson L (2012) Development of marker sets useful in the early selection of Ren4 powdery mildew resistance and seedlessness for table and raisin grape breeding. Theor Appl Genet 124:23–33
Martins WS, Lucas DC, Neves KF, Bertioli DJ (2009) WebSat—a web software for microsatellite marker development. Bioinformation 3:282–283
Moroldo M, Paillard S, Marconi R, Fabrice L, Canaguier A, Cruaud C, De Berardinis V, Guichard C, Brunaud V, Le Clainche I, Scalabrin S, Testolin R, Di Gaspero G, Morgante M, Adam-Blondon AF (2008) A physical map of the heterozygous grapevine ‘Cabernet Sauvignon’ allows mapping candidate genes for disease resistance. BMC Plant Biol 8:66
Myles S, Chia J-M, Hurwitz B, Simon C, Zhong G-Y, Buckler E, Ware D (2010) Rapid genomic characterization of the genus Vitis. PLoS One 5:e8219
Ramming DW, Gabler F, Smilanick J, Cadle-Davidson M, Barba P, Mahanil S, Cadle-Davidson L (2011) A single dominant locus, Ren4, confers rapid non-race-specific resistance to grapevine powdery mildew. Phytopathology 101:502–508
Rex F, Fechter I, Hausmann L, Topfer R (2014) QTL mapping of black rot (Guignardia bidwellii) resistance in the grapevine rootstock ‘Börner’ (V. riparia Gm183× V. cinerea Arnold). Theor Appl Genet 127:1667–1677
Riaz S, Tenscher AC, Ramming DW, Walker MA (2011) Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor Appl Genet 122:1059–1073
Sadras VO, Petrie PR (2011) Climate shifts in south-eastern Australia: early maturity of Chardonnay, Shiraz and Cabernet Sauvignon is associated with early onset rather than faster ripening. Aust J Grape Wine Res 17:199–205
Schwander F, Eibach R, Fechter I, Hausmann L, Zyprian E, Töpfer R (2012) Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Genet 124:163–176
Tena G, Boudsocq M, Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14:519–529
van Heerden CJ, Burger P, Vermeulen A, Prins R (2014) Detection of downy and powdery mildew resistance QTL in a ‘Regent’ × ‘Red Globe’ population. Euphytica 200:281–295
Van Ooijen JW (2006) JoinMap®4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Van Ooijen JW (2009) MapQTL6®, Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B.V., Wageningen
Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma T, Facci M, Mitchell JT, Perazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Dematte L, Mraz A, Battilana J, Stormo K, Costa F, Tao QZ, Si-Ammour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett JA, Sterck L, Vandepoele K, Grando SM, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaramella V, Bhatnagar SK, Fontana P, Gutin A, Van de Peer Y, Salamini F, Viola R (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2(12):e1326
Venuti S, Copetti D, Foria S, Falginella L, Hoffmann S, Bellin D, Cindric P, Kozma P, Scalabrin S, Morgante M, Testolin R, Di Gaspero G (2013) Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS One 8(4):e61228
Welter LJ, Göktürk-Baydar N, Akkurt M, Maul E, Eibach R, Töpfer R, Zyprian EM (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol Breeding 20:359–374
Xiao H, Tattersall EAR, Siddiqua MK, Cramer GR, Nassuth A (2008) CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant, Cell Environ 31:1–10
Zhang JK, Hausmann L, Eibach R, Welter LJ, Töpfer R, Zyprian EM (2009) A framework map from grapevine V3125 (Vitis vinifera ‘Schiava grossa’ x ‘Riesling’) x rootstock cultivar ‘Börner’ (Vitis riparia x Vitis cinerea) to localize genetic determinants of phylloxera root resistance. Theor Appl Genet 119:1039–1051
Zyprian E (unpublished) QTL analysis of flowering and veraison time
Zyprian E, Eibach R, Töpfer R (2005) Eine neue genetische Karte der Weinrebe aus der Kreuzung “Gf.Ga-47-42 × ”Villard blanc”. Deutsches Weinbau Jahrbuch 2006 Verlag Eugen Ulmer 57:151–158
Acknowledgments
We thank Doreen Gabriel for help with statistical analyses. We also thank Elvira Schreiber for assistance with the analysis of flowering and veraison time. Heike Bennek, Margit Schneider, and Andreas Preiss provided expert technical help with marker analyses.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the German Federal Ministry of Nutrition and Agriculture and partially by InnoVine (grant 311775). MBR was supported by ERA-Net (through the German Federal Ministry of Education and Research) and SS received a fellowship from L’Organisation Internationale de la Vigne et du Vin. FS and IO received funding from Forschungsring des Deutschen Weinbaues and Deutsche Forschungsgemeinschaft (SPP1530), respectively.
Conflict of interest
All authors have no conflicts of interest to declare.
Ethical standards
The authors declare that all experiments conducted in this study complied with the applicable laws in Germany and France. This article does not describe any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 1
Distribution of veraison time variation (a) Continuous distribution of the values for the timing of the onset of veraison in the segregating population in 2010. The complete range extended over 45 days. Veraison occurred in GF.GA-47-42 and ‘Villard blanc’ lines 58 and 91 days after June 1, respectively. (b) Frequency distribution of the timing of the onset of veraison in the segregating population in 2010. (PDF 57 kb)
Table 1
(a) Designations, chromosome positions, sequences, and amplimer sizes of newly designed SSR markers.(PDF 48 kb)
Table 1
(b) Designations, chromosome positions, and biallelic nucleotide polymorphisms of novel SNP markers. (PDF 34 kb)
Table 2
Parameters used to evaluate Plasmopara viticola resistance on leaf discs(PDF 101 kb)
Table 3
Segregation patterns and genetic information gained from the three sets of SNP markers. (PDF 35 kb)
Table 4
(a) Spearman rank correlation coefficients of Plasmopara viticola resistance traits for leaves and berries over 3 years in the field and for leaf discs under in vitro culture conditions. (PDF 180 kb)
Table 4
(b) Spearman rank correlation coefficients of Erysiphe necator resistance traits for leaves and berries over 6 years in the field. (PDF 191 kb)
Table 4
(c) Spearman rank correlation coefficients of the time required to reach the flowering and veraison stages, and the interval between the two developmental stages over 3 or 4 years. (PDF 98 kb)
Rights and permissions
About this article
Cite this article
Zyprian, E., Ochßner, I., Schwander, F. et al. Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol Genet Genomics 291, 1573–1594 (2016). https://doi.org/10.1007/s00438-016-1200-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1200-5