Abstract
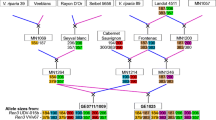
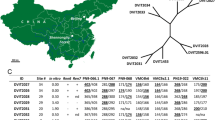
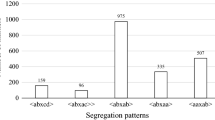
The single, dominant powdery mildew resistance locus Ren4 from Vitis romanetii prevents hyphal growth by Erysiphe necator. Previously, we showed that when introgressed into V. vinifera in the modified BC2 population 03-3004, Ren4 was linked with the simple sequence repeat marker VMC7f2 on chromosome 18—a marker that is associated with multiple disease resistance and seedlessness. However, in the current study, this marker was monomorphic in related breeding populations 05-3010 and 07-3553. To enhance marker-assisted selection at this locus, we developed multiplexed SNP markers using three approaches: conversion of bulked segregant analysis AFLP markers, sequencing of candidate genes and regions flanking known V. vinifera SNPs, and hybridization to the Vitis9KSNP genotyping array. The Vitis9KSNP array was more cost-efficient than all other approaches tested for marker discovery and genotyping, enabling the genotyping of 1317 informative SNPs within the span of 1 week and at a cost of 11 cents per SNP. From a total of 1,446 high quality, informative markers segregating in 03-3004, we developed a haplotype signature of 15 multiplexed SNP markers linked with Ren4 in 03-3004, 5 of which were linked in 05-3010, and 6 of which were linked in 07-3553. Two of these populations segregated for seedlessness, which was tightly linked with Ren4 in 03-3004 (2 cM) but not in 05-3010 (22 cM). Chromosomal rearrangements were detected among these three populations and the reference genome PN40024. Since this is the first application of the Vitis9KSNP array in a breeding program, some suggestions are provided for application of genotyping arrays. Our results provide novel markers for tracking and pyramiding this unique resistance gene and for further functional characterization of this region on chromosome 18 encoding multiple disease resistance and seedlessness.



Similar content being viewed by others
References
Adam-Blondon A-F, Roux C, Claux D, Butterlin G, Merdinoglu D, This P (2004) Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet 109:1017–1027
Akkurt M, Welter L, Maul E, Topfer R, Zyprian E (2007) Development of SCAR markers linked to powdery mildew (Uncinula necator) resistance in grapevine (Vitis vinifera L. and Vitis sp.). Mol Breed 19(2):103–111
Arroyo-Garcia R, Martinez-Zapater JM (2004) Development and characterization of new microsatellite markers for grape. Vitis 4:175–178
Barker CL, Donald T, Pauquet J, Ratnaparkhe MB, Bouquet A, Adam-Blondon AF, Thomas MR, Dry I (2005) Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor Appl Genet 111(2):370–377
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon AF, Cipriani G, Morgante M, Testolin R, Di Gaspero G (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet 120(1):163–176
Bowers JE, Dangl GS, Meredith CP (1999) Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic 50:243–246
Bowers JE, Dangl GS, Vignani R, Meredith CP (1996) Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 39:628–633
Cabezas JA, Cervera MT, Ruiz-Garcia L, Carreno J, Martinez-Zapater JM (2006) A genetic analysis of seed and berry weight in grapevine. Genome 49:1572–1585
Cadle-Davidson L, Chicoine DR, Consolie NC (2011) Variation within and between Vitis species for foliar resistance to the powdery mildew pathogen Erysiphe necator. Plant Dis 95:202–211
Coleman C, Copetti D, Cipriani G, Hoffman S, Kozma P, Kovacs L, Morgante M, Testolin R, Di Gaspero G (2009) The powdery mildew resistance gene REN1 co-segregates with an NBS-LRR gene cluster in two Central Asian grapevines. BMC Genet 10:89
Costantini L, Grando MS, Feingold S, Ulanovsky S, Mejia N, Hinrichsen P, Doligez A, This P, Cabezas JA, Martinez-Zapater JM (2007) Generation of a common set of mapping markers to assist table grape breeding. Am J Enol Vitic 58(1):102–111
Dalbo MA, Ye GN, Weeden NF, Wilcox WF, Reisch BI (2001) Marker-assisted selection for powdery mildew resistance in grapes. J Am Soc Hortic Sci 126(1):83–89
Di Gaspero G, Peterlunger E, Testolin R, Edwards KJ, Cipriani C (2000) Conservation of microsatellite loci within the genus Vitis. Theor Appl Genet 101:301–308
Donald TM, Pellerone F, Adam-Blondon AF, Bouquet A, Thomas MR, Dry IB (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 104:610–618
Eibach R, Zyprian E, Welter L, Topfer R (2007) The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis 46(3):120–124
Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Topfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108(3):501–515
Gorg R, Hollricher K, Schulze-Lefert P (1993) Functional analysis and RFLP-mediated mapping of the Mlg resistance locus in barley. Plant J 3(6):857–866
Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, Maroof MAS (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166(1):493–503
Jorgensen JH (1992) Discovery, characterization and exploitation of mlo powdery mildew resistance in barley. Euphytica 63(1–2):141–152
Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16(11):2870–2894
Mahanil S, Reisch BI, Owens CL, Thipyapong P, Laosuwan P (2007) Resistance gene analogs from Vitis cinerea, Vitis rupestris, and Vitis hybrid Horizon. Am J Enol Vitic 58(4):484–493
Mahanil S, Lagerholm S, Cadle-Davidson L (2009) Pattern of genetic variation in Mlo among Vitis spp. and interspecific hybrids. IS-MPMI 2009
Mahanil S, Lagerholm S, Garris A, Owens CL, Ramming DW, Cadle-Davidson L (2011) Development of molecular markers for powdery mildew resistance in grapevines. Acta Hortic (in press)
Marano MR, Malcuit I, De Jong W, Baulcombe DC (2002) High-resolution genetic map of Nb, a gene that confers hypersensitive resistance to potato virus X in Solanum tuberosum. Theor Appl Genet 105(2–3):192–200
Mejia N, Gebauer M, Munoz L, Hewstone N, Munoz C, Hinrichsen P (2007) Identification of QTLs for seedlessness, berry size, and ripening date in a seedless × seedless table grape progeny. Am J Enol Vitic 58(4):499–507
Merdinoglu D, Butterlin G, Bevilacqua L, Chiquet V, Adam-Blondon A-F, Decrooocq S (2005) Development and characterization of a large set of microsattellite markers in grapevine (Vitis vinifera L.) suitable for mutiplex PCR. Mol Breed 15:349–366
Menzel S, Qin J, Vasavda N, Thein SL, Ramakrishnan R (2010) Experimental generation of SNP haplotype signatures in patients with sickle cell anaemia. PLoS One 5(9):e13004
Moroldo M, Paillard S, Marconi R, Fabrice L, Canaguier A, Cruaud C, De Berardinis V, Guichard C, Brunaud V, Le Clainche I, Scalabrin S, Testolin R, Di Gaspero G, Morgante M, Adam-Blondon AF (2008) A physical map of the heterozygous grapevine ‘Cabernet Sauvignon’ allows mapping candidate genes for disease resistance. BMC Plant Biol 8:66
Myles S, Chia JM, Hurwitz B, Simon C, Zhong GY, Buckler E, Ware D (2010) Rapid genomic characterization of the genus Vitis. PLoS One 5(1):9
Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, Prins B, Reynolds A, Chia J-M, Ware D, Bustamante CD, Buckler ES (2011) Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA 108:3530–3535
Pauquet J, Bouquet A, This P, Adam-Blondon AF (2001) Establishment of a local map of AFLP markers around the powdery mildew resistance gene Run1 in grapevine and assessment of their usefulness for marker assisted selection. Theor Appl Genet 103:1201–1210
Peressotti E, Wiedemann-Merdinoglu S, Delmotte F, Bellin D, Di Gaspero G, Testolin R, Merdinoglu D, Mestre P (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol 10:147
Ramming DW (1990) The use of embryo culture in fruit breeding. Hortscience 25(4):393–398
Ramming DW, Gabler F, Smilanick J, Cadle-Davidson M, Barba P, Mahanil S, Cadle-Davidson L (2011) A single dominant locus Ren4 confers non-race-specific penetration resistance to grapevine powdery mildew. Phytopathology 101:502–508
Riaz S, Tenscher AC, Ramming DW, Walker MA (2011) Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor Appl Genet 122:1059–1073
Sefc KM, Regner F, Turetschek E, Gloss J, Steinkellner H (1999) Identification of microsattelite sequence in Vitis species. Genome 42:367–373
Troggio M, Malacarne G, Coppola G, Segala C, Cartwright DA, Pindo M, Stefanini M, Mank R, Moroldo M, Morgante M, Grando MS, Velasco R (2007) A dense single-nucleotide polymorphism-based genetic linkage map of grapevine (Vitis vinifera L.) anchoring pinot noir bacterial artificial chromosome contigs. Genetics 176:2637–2650
van der Voort JR, Kanyuka K, van der Vossen E, Bendahmane A, Mooijman P, Klein-Lankhorst R, Stiekema W, Baulcombe D, Bakker J (1999) Tight physical linkage of the nematode resistance gene Gpa2 and the virus resistance gene Rx on a single segment introgressed from the wild species Solanum tuberosum subsp andigena CPC1673 into cultivated potato. Mol Plant Microbe Interact 12(3):197–206
Van Ooijen JW (2006) JoinMap®4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Van Ooijen JW (2009) MapQTL® 6, software for the mapping and quantitative trait loci in experimental population of diploid species. Kyazma B.V., Wageningen
Vezzulli S, Micheletti D, Riaz S, Pindo M, Viola R, This P, Walker MA, Troggio M, Velasco R (2008) A SNP transferability survey within the genus Vitis. BMC Plant Biol 8:128
Wan Y, Schwaninger H, He P, Wang Y (2007) Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 46(3):132–136
Wang W, Wen Y, Berkey R, Xiao S (2009) Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21:2898–2913
Welter LJ, Gokturk-Baydar N, Akkurt M, Maul E, Eibach R, Topfer R, Zyprian EM (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L.). Mol Breed 20:359–374
Zhang ZG, Feechan A, Pedersen C, Newman MA, Qiu JL, Olesen KL, Thordal-Christensen H (2007) A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J 49(2):302–312
Acknowledgments
This project was funded by the American Vineyard Foundation. Development of the Vitis9KSNP array and its application in genotyping of the parents and 18 progeny was funded by a grant from USDA-ARS to Ed Buckler and Doreen Ware.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
S. Mahanil and D. Ramming equally contributed to this study.
Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mahanil, S., Ramming, D., Cadle-Davidson, M. et al. Development of marker sets useful in the early selection of Ren4 powdery mildew resistance and seedlessness for table and raisin grape breeding. Theor Appl Genet 124, 23–33 (2012). https://doi.org/10.1007/s00122-011-1684-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1684-7