Abstract
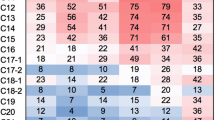
Microbial community exhibit shift in composition in response to temperature variation. We report crude oil-degrading activity and high-throughput 16S rRNA gene sequencing (metagenome) profiles of four bacterial consortia enriched at three different temperatures in crude oil-amended Bushnell–Hass Medium from an oily sludge sediment. The consortia were referred to as O (4 ± 2 ℃ in 3% w/v crude oil), A (25 ± 2 ℃ in 1% w/v crude oil), H (25 ± 2 ℃ in 3% w/v crude oil), and X (45 ± 2 ℃ in 3% w/v crude oil). The hydrocarbon-degrading activity was highest for consortium A and H and lowest for consortium O. The metagenome profile revealed the predominance of Proteobacteria (62.12–1.25%) in each consortium, followed by Bacteroidota (18.94–37.77%) in the consortium O, A, and H. Contrarily, consortium X comprised 7.38% Actinomycetota, which was essentially low (< 0.09%) in other consortia, and only 0.41% Bacteroidota. The PICRUSt-based functional analysis predicted major functions associated with the metabolism and 5060 common KEGG Orthology (KOs). A total of 296 KOs were predicted exclusively in consortium X. Additionally, 247 KOs were predicted from xenobiotic biodegradation pathways. This study found that temperature had a stronger influence on the composition and function of the bacterial community than crude oil concentration.






Similar content being viewed by others
References
Sexstone A, Everett K, Jenkins T, Atlas RM (1978) Fate of crude and refined oils in north slope soils. Arctic 31(3):339–347
Zhang Y, Dong S, Wang H, Tao S, Kiyama R (2016) Biological impact of environmental polycyclic aromatic hydrocarbons (ePAHs) as endocrine disruptors. Environ Pollut 213:809–824
Bashir I, Lone FA, Bhat RA, Mir SA, Dar ZA, Dar SA (2020) Concerns and threats of contamination on aquatic ecosystems. Bioremediation and Biotechnology: Sustainable Approaches to Pollution Degradation. Springer International Publishing, Cham, pp 1–26
Singh AK, Sherry A, Gray ND, Jones DM, Bowler BF, Head IM (2014) Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front Microbiol 5:160
Chettri B, Mukherjee A, Langpoklakpam JS, Chattopadhyay D, Singh AK (2016) Kinetics of nutrient enhanced crude oil degradation by Pseudomonas aeruginosa AKS1 and Bacillus sp. AKS2 isolated from Guwahati refinery. India Environ Pollut 216:548–558
Nedwell DB (1999) Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiol Ecol 30:101–111
Bissett A, Richardson AE, Baker G, Wakelin S, Thrall PH (2010) Life history determines biogeographical patterns of soil bacterial communities over multiple spatial scales. Mol Ecol 19(19):4315–4327
Northcott GL, Jones KC (2000) Experimental approaches and analytical techniques for determining organic compound bound residues in soil and sediment. Environ Pollut 108:19–43
Rowland AP, Lindley DK, Hall GH, Rossall MJ, Wilson DR, Benham DG, Harrison AF, Daniels RE (2000) Effects of beach sand properties, temperature and rainfall on the degradation rates of oil in buried oil/beach sand mixtures. Environ Pollut 109:109–118
Whyte LG, Hawari J, Zhou E, Bourbonnière L, Inniss WE, Greer CW (1998) Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl Environ Microbiol 64:2578–2584
Rojo F (2009) Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490
Mohn WW, Stewart GR (2000) Limiting factors for hydrocarbon biodegradation at low temperature in Arctic soils. Soil Biol Biochem 32(8–9):1161–1172
Sikkema J, de Bont JA, Poolman B (1995) Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59(2):201–222
Delille D, Coulon F, Pelletier E (2004) Effects of temperature warming during a bioremediation study of natural and nutrient-amended hydrocarbon-contaminated sub-Antarctic soils Cold Region. Sci Technol 40:61–70
Coulon F, Pelletier E, Gourhant L, Delille D (2005) Effects of nutrient and temperature on degradation of petroleum hydrocarbons in contaminated sub-Antarctic soil. Chemosphere 58:1439–1448
Margesin R, Neuner G, Storey K (2007) Cold-loving microbes, plants, and animals – fundamental and applied aspects. Naturwissenschaften 94:77–99
Yergeau E, Kowalchuk GA (2008) Responses of Antarctic soil microbial communities and associated functions to temperature and freeze-thaw cycle frequency. Environ Microbial 10(9):2223–2235
Yergeau E, Sanschagrin S, Beaumier D, Greer CW (2012) Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS One 7(1):30058
Mukherjee A, Chettri B, Langpoklakpam JS, Basak P, Prasad A, Mukherjee AK, Bhattacharyya M, Singh AK, Chattopadhyay D (2017) Bioinformatic approaches including predictive metagenomic profiling reveal characteristics of bacterial response to petroleum hydrocarbon contamination in diverse environments. Sci Rep 7(1):1–22
Roy A, Dutta A, Pal S, Gupta A, Sarkar J, Chatterjee A, Saha A, Sarkar P, Sar P, Kazy SK (2018) Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Biores Technol 253:22–32
Devi SP, Jani K, Sharma A, Jha DK (2022) Bacterial communities and their bioremediation capabilities in oil-contaminated agricultural soils. Environ Monit Assess 194:1–13
Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30(15):2114–2120
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol 31(9):814–821
Chen W, Kong Y, Li J, Sun Y, Min J, Hu X (2020) Enhanced biodegradation of crude oil by constructed bacterial consortium comprising salt-tolerant petroleum degraders and biosurfactant producers. Int Biodeter Biodegrad 154:105047
Varjani S, Upasani VN (2019) Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil. J Environ Manage 1(245):358–366
Suja F, Rahim F, Taha MR, Hambali N, Razali MR, Khalid A, Hamzah A (2014) Effects of local microbial bioaugmentation and biostimulation on the bioremediation of total petroleum hydrocarbons (TPH) in crude oil contaminated soil based on laboratory and field observations. Int Biodeter Biodegrad 90:115–122
Baltaci MO, Omeroglu MA, Ozkan H, Taskin M Adıguzel A (2023) Enhanced biodegradation of crude oil contamination by indigenous bacterial consortium under real conditions. Biocatal Biotransfor 1–12.
Nnabuife OO, Ogbonna JC, Anyanwu C, Ike AC, Eze CN, Enemuor SC (2022) Mixed bacterial consortium can hamper the efficient degradation of crude oil hydrocarbons. Arch Microbiol 204(6):306
Yang Y, Wang J, Liao J, Xie S, Huang Y (2014) Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Appl Microbiol Biotech 99:1935–1946
Peng M, Zi X, Wang Q (2015) Bacterial community diversity of oil-contaminated soils assessed by high throughput sequencing of 16S rRNA genes. Int J Environ Res Public Health 12:12002–12015
Wu M, Ye X, Chen K, Li W, Yuan J, Jiang X (2017) Bacterial community shift and hydrocarbon transformation during bioremediation of short-term petroleum-contaminated soil. Environ Pollut 223:657–664
Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M (2011) Hydrocarbon degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater horizon oil spill. Appl Environ Microbiol 77(22):7962–7974
Pi YR, Bao MT (2022) Investigation of kinetics in bioaugmentation of crude oil via high-throughput sequencing: enzymatic activities, bacterial community composition and functions. Petrol Sci 19(4):1905–1914
D’Ugo E, Bruno M, Mukherjee A, Chattopadhyay D, Giuseppetti R, De Pace R, Magurano F (2021) Characterization of microbial response to petroleum hydrocarbon contamination in a lacustrine ecosystem. Environ Sci Pollut Res 28(20):26187–26196
Sarkar J, Saha A, Roy A, Bose H, Pal S, Sar P, Kazy SK (2020) Development of nitrate stimulated hydrocarbon-degrading microbial consortia from refinery sludge as potent bioaugmenting agent for enhanced bioremediation of petroleum contaminated waste. World J Microbiol Biotechnol 36(10):1–20
Qiu Y, Xiao F, Wei X, Wen Z, Chen S (2014) Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol 98(21):8895–8903
Medic A, Ljesevic M, Inui H, Beskoski V, Kojic I, Stojanovic K, Karadzic I (2020) Efficient biodegradation of petroleum n-alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity. RSC Adv 10:14060–14070
Chettri B, Singha NA, Singh AK (2021) Efficiency and kinetics of Assam crude oil degradation by Pseudomonas aeruginosa and Bacillus sp. Arch Microbiol 203(9):5793–5803
Das R, Kazy SK (2014) Microbial diversity, community composition and metabolic potential in hydrocarbon contaminated oily sludge: prospects for in situ bioremediation. Environ Sci Pollut Res 21(12):7369–7389
Silva TR, Verde LCL, Neto ES, Oliveira VM (2013) Diversity analyses of microbial communities in petroleum samples from Brazilian oil fields. Int Biodeter Biodegrad 81:57–70
Chaudhary DK, Kim DU, Kim D, Kim J (2019) Flavobacterium petrolei sp. nov. a novel psychrophilic, diesel-degrading bacterium isolated from oil-contaminated Arctic soil. Sci Rep 9(1):1–9
Liu B, Yang X, Sheng M, Yang Z, Qiu J, Wang C, He J (2020) Sphingobacterium olei .sp nov., isolated from oil-contaminated soil. Int J Systemat Evolut Microbiol 70(3):1931–1939
Margesin R, Spröer C, Schumann P, Schinner F (2003) Pedobacter cryoconitis sp. nov., a facultative psychrophile from alpine glacier cryoconite. Int J Systemat Evolut Microbiol 53(5):1291–1296
Gao P, Tian H, Wang Y, Li Y, Li Y, Xie J, Zeng B, Zhou J, Li G, Ma T (2016) Spatial isolation and environmental factors drive distinct bacterial and archaeal communities in different types of petroleum reservoirs in China. Sci Rep 6(1):1–12
Pannekens M, Kroll L, Müller H, Mbow FT, Meckenstock RU (2019) Oil reservoirs, an exceptional habitat for microorganisms. New Biotechnol 49:1–9
Neihsial R, Singha NA, Singh AK (2022) Taxonomic diversity and predictive metabolic functions of a heavy metal tolerant multiple azo dye degrading bacterial consortium from textile effluents. Inter Biodeter Biodegr 171:105421
Omrani R, Spini G, Puglisi E, Saidane D (2018) Modulation of microbial consortia enriched from different polluted environments during petroleum biodegradation. Biodegradation 29(2):187–209
Kappell AD, Wei Y, Newton RJ, Van Nostrand JD, Zhou J, McLellan SL, Hristova KR (2014) The polycyclic aromatic hydrocarbon degradation potential of Gulf of Mexico native coastal microbial communities after the Deepwater Horizon oil spill. Front Microbiol 5:205
Chikere CB, Mordi IJ, Chikere BO, Selvarajan R, Ashafa TO, Obieze CC (2019) Comparative metagenomics and functional profiling of crude oil-polluted soils in Bodo West Community, Ogoni, with other sites of varying pollution history. Ann Microbiol 69(5):495–513
Bao YJ, Xu Z, Li Y, Yao Z, Sun J, Song H (2017) High-throughput metagenomic analysis of petroleum-contaminated soil microbiome reveals the versatility in xenobiotic aromatics metabolism. J Environ Sci (China) 56:25–35
Acknowledgements
This work was supported by a Department of Science and Technology, Government of India grant number: CRG/2022/002550. We are thankful to the Head, Department of Biochemistry, North-Eastern Hill University, India for providing the infrastructural facility for carrying out this research study. Fellowship grant from DRS-III SAP, UGC to NAS, and DR DS Kothari Fellowship to PS is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
AKS and NAS conceptualized and designed the experiments. NAS, RN, BC, and WJL conducted experiments for biodegradation of crude oil. NAS and RN designed and conducted the bioinformatics analysis strategy with assistance from LK, WJL, JN, and PS for data analyses. AKS and NAS prepared the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singha, N.A., Neihsial, R., Kipgen, L. et al. Taxonomic and Predictive Functional Profile of Hydrocarbonoclastic Bacterial Consortia Developed at Three Different Temperatures. Curr Microbiol 81, 22 (2024). https://doi.org/10.1007/s00284-023-03529-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03529-0