Abstract
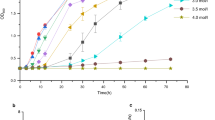
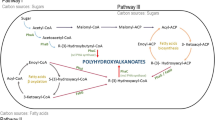
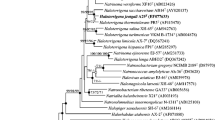
Species of the Halomonas genus are gram-negative, aerobic, moderately halophilic bacteria that synthesize polyhydroxyalkanoates (PHAs) and other high-value products that have a wide range of potential uses in the food, feed, cosmetics, pharmaceutical, and chemical sectors. Genome sequencing studies allow for the description and comparison of genetic traits with other strains and species, allowing for the exploration of the organism's potential, necessary to further biotechnology applications. Here, the genome of Halomonas elongata strain 153B was sequenced, its features compared to 5 other strains and 7 species, and a description of features for adaptations to hypersaline environments and bioproducts synthesis was done. Whole-genome analysis showed H. elongata 153B has more similar features to the reference strain H. elongata DSM 2581 compared to 4 other reported strains. Comparative genomics showed 2064 core genomic clusters between the strains and 666 singletons for strain 153B. Several genes in transport and signaling, osmoregulation, and oxidative stress that have roles in adaptation to environments with high osmolarity were also revealed. These appear to form an intricate network of overlapping systems carefully coordinated to bring about adaptation. H. elongata 153B genes for the synthesis of PHAs, ectoine, vitamins, and the degradation of drugs and aromatic compounds were described. The results will aid in the study of halophile physiology, provide a mine for valuable enzymes, and help speed up research for other biotechnology applications.





Similar content being viewed by others
Data Availability
The genomic sequence data in this study were deposited in the NCBI database.
Code Availability
Not applicable.
References
Arahal DR, Ventosa A (2006) The family halomonadaceae. The family halomonadaceae. Springer, New York
Daoud L, Ben Ali M (2020) Chapter 5—Halophilic microorganisms: Interesting group of extremophiles with important applications in biotechnology and environment. In: Salwan R, Sharma V (eds) Physiological and biotechnological aspects of extremophiles. Academic Press, pp 51–64
Çakmak H, Çelik PA, Çınar S et al (2021) Levan production potentials from different hypersaline environments in Turkey. J Microbiol, Biotechnol Food Sci. https://doi.org/10.15414/jmbfs.2020.10.1.61-64
Radchenkova N, Hasköylü ME, Vassilev S et al (2020) Improved exopolymer production by Chromohalobacter canadensis cultures for its potential cosmeceutical applications. Microorganisms 8:1935. https://doi.org/10.3390/microorganisms8121935
Ye J-W, Chen G-Q (2021) Halomonas as a chassis. Essays Biochem 65:393–403. https://doi.org/10.1042/EBC20200159
Qin Q, Ling C, Zhao Y et al (2018) CRISPR/Cas9 editing genome of extremophile Halomonas spp. Metab Eng 47:219–229. https://doi.org/10.1016/j.ymben.2018.03.018
Jiang X-R, Yao Z-H, Chen G-Q (2017) Controlling cell volume for efficient PHB production by Halomonas. Metab Eng 44:30–37. https://doi.org/10.1016/j.ymben.2017.09.004
Ma H, Zhao Y, Huang W et al (2020) Rational flux-tuning of Halomonas bluephagenesis for co-production of bioplastic PHB and ectoine. Nat Commun 11:3313. https://doi.org/10.1038/s41467-020-17223-3
Thomas T, Elain A, Bazire A, Bruzaud S (2019) Complete genome sequence of the halophilic PHA-producing bacterium Halomonas sp. SF2003: insights into its biotechnological potential. World J Microbiol Biotechnol 35:50. https://doi.org/10.1007/s11274-019-2627-8
Mukherjee S, Stamatis D, Bertsch J et al (2019) Genomes OnLine database (GOLD) vol 7: updates and new features. Nucleic Acids Res 47:D649–D659. https://doi.org/10.1093/nar/gky977
Ye J, Hu D, Che X et al (2018) Engineering of Halomonas bluephagenesis for low cost production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) from glucose. Metab Eng 47:143–152. https://doi.org/10.1016/j.ymben.2018.03.013
Enuh BM, Nural Yaman B, Tarzi C et al (2022) Whole-genome sequencing and genomescale metabolic modeling of Chromohalobacter canadensis 85B to explore its salt tolerance and biotechnological use. MicrobiologyOpen. 11:e1328. https://doi.org/10.1002/mbo3.1328
Çınar S, Mutlu MB (2016) Comparative analysis of prokaryotic diversity in solar salterns in eastern Anatolia (Turkey). Extremophiles 20:589–601. https://doi.org/10.1007/s00792-016-0845-7
Gedikli S, Aytar Çelik P, Demirbilek M et al (2019) Experimental exploration of thermostable poly (β-hydroxybutyrates) by Geobacillus kaustophilus using Box-Behnken design. J Polym Environ. https://doi.org/10.1007/s10924-018-1335-z
Wattam AR, Brettin T, Davis JJ et al (2018) Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol Biol 1704:79–101. https://doi.org/10.1007/978-1-4939-7463-4_4
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. https://doi.org/10.1371/journal.pcbi.1005595
Brettin T, Davis JJ, Disz T et al (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. https://doi.org/10.1038/srep08365
Boutet E, Lieberherr D, Tognolli M et al (2007) UniProtKB/Swiss-Prot. Methods Mol Biol 406:89–112. https://doi.org/10.1007/978-1-59745-535-0_4
Moriya Y, Itoh M, Okuda S et al (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. https://doi.org/10.1093/nar/gkm321
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Huerta-Cepas J, Szklarczyk D, Heller D et al (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. https://doi.org/10.1093/nar/gky1085
Stothard P, Grant JR, Van Domselaar G (2019) Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform 20:1576–1582. https://doi.org/10.1093/bib/bbx081
Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. https://doi.org/10.1186/1471-2164-12-402
Xu L, Dong Z, Fang L et al (2019) OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 47:W52–W58. https://doi.org/10.1093/nar/gkz333
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.2307/2408678
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford, New York
Wang Q, Nie P, Hou Y, Wang Y (2020) Purification, biochemical characterization and DNA protection against oxidative damage of a novel recombinant superoxide dismutase from psychrophilic bacterium Halomonas sp ANT108. Protein Expr Purif 173:105661. https://doi.org/10.1016/j.pep.2020.105661
Diken E, Ozer T, Arikan M et al (2015) Genomic analysis reveals the biotechnological and industrial potential of levan producing halophilic extremophile, Halomonas smyrnensis AAD6T. Springerplus 4:393. https://doi.org/10.1186/s40064-015-1184-3
Alm E, Huang K, Arkin A (2006) The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2:e143. https://doi.org/10.1371/journal.pcbi.0020143
Grammann K, Volke A, Kunte HJ (2002) New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581T. J Bacteriol. https://doi.org/10.1128/JB.184.11.3078-3085.2002
Poli A, Nicolaus B, Denizci AA et al (2013) Halomonas smyrnensis sp. nov., a moderately halophilic, exopolysaccharide-producing bacterium. Int J Syst Evol Microbiol 63:10–18. https://doi.org/10.1099/ijs.0.037036-0
Gunde-Cimerman N, Plemenitaš A, Oren A (2018) Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev 42:353–375. https://doi.org/10.1093/femsre/fuy009
Kraegeloh A, Amendt B, Kunte HJ (2005) Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T. J Bacteriol 187:1036–1043. https://doi.org/10.1128/JB.187.3.1036-1043.2005
Chen Y-H, Lu C-W, Shyu Y-T, Lin S-S (2017) Revealing the saline adaptation strategies of the halophilic bacterium Halomonas beimenensis through high-throughput omics and transposon mutagenesis approaches. Sci Rep 7:13037. https://doi.org/10.1038/s41598-017-13450-9
Chen Y-H, Shyu Y-T, Lin S-S (2018) Characterization of candidate genes involved in halotolerance using high-throughput omics in the halotolerant bacterium Virgibacillus chiguensis. PLoS ONE 13:e0201346. https://doi.org/10.1371/journal.pone.0201346
Ongagna-Yhombi SY, McDonald ND, Boyd EF (2015) Deciphering the role of multiple betaine-carnitine-choline transporters in the halophile Vibrio parahaemolyticus. Appl Environ Microbiol 81:351–363. https://doi.org/10.1128/AEM.02402-14
Cummings SP, Gilmour DJ (1995) The effect of NaCl on the growth of a halomonas species: accumulation and utilization of compatible solutes. Microbiology 141:1413–1418. https://doi.org/10.1099/13500872-141-6-1413
Nakayama H, Yoshida K, Ono H et al (2000) Ectoine, the compatible solute of Halomonas elongata, confers hyperosmotic tolerance in cultured tobacco cells. Plant Physiol 122:1239–1247. https://doi.org/10.1104/pp.122.4.1239
Calamita G (2000) The Escherichia coli aquaporin-Z water channel. Mol Microbiol 37:254–262. https://doi.org/10.1046/j.1365-2958.2000.02016.x
Lang S, Cressatti M, Mendoza KE et al (2015) YehZYXW of Escherichia coli Is a low-affinity, non-osmoregulatory betaine-specific ABC transporter. Biochemistry 54:5735–5747. https://doi.org/10.1021/acs.biochem.5b00274
Gostinčar C, Gunde-Cimerman N (2018) Overview of oxidative stress response genes in selected halophilic fungi. Genes (Basel) 9:E143. https://doi.org/10.3390/genes9030143
Sztukowska M, Bugno M, Potempa J et al (2002) Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol 44:479–488. https://doi.org/10.1046/j.1365-2958.2002.02892.x
Zheng Y, Chen J-C, Ma Y-M, Chen G-Q (2020) Engineering biosynthesis of polyhydroxyalkanoates (PHA) for diversity and cost reduction. Metab Eng 58:82–93. https://doi.org/10.1016/j.ymben.2019.07.004
Chek MF, Kim S-Y, Mori T et al (2017) Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp USM2, producing biodegradable plastics. Sci Rep 7:5312. https://doi.org/10.1038/s41598-017-05509-4
McCool GJ, Cannon MC (2001) PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J Bacteriol 183:4235–4243. https://doi.org/10.1128/JB.183.14.4235-4243.2001
Velázquez-Sánchez C, Espín G, Peña C, Segura D (2020) The modification of regulatory circuits involved in the control of polyhydroxyalkanoates metabolism to improve their production. Front Bioeng Biotechnol 8:386. https://doi.org/10.3389/fbioe.2020.00386
Choi MH, Xu J, Gutierrez M et al (2011) Metabolic relationship between polyhydroxyalkanoic acid and rhamnolipid synthesis in Pseudomonas aeruginosa: comparative 13C NMR analysis of the products in wild-type and mutants. J Biotechnol 151:30–42. https://doi.org/10.1016/j.jbiotec.2010.10.072
Hoffmann N, Rehm BHA (2004) Regulation of polyhydroxyalkanoate biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa. FEMS Microbiol Lett 237:1–7. https://doi.org/10.1016/j.femsle.2004.06.029
de Moreno ML, Sánchez-Porro C, Piubeli F et al (2011) Cloning, characterization and analysis of cat and ben genes from the phenol degrading halophilic bacterium Halomonas organivorans. PLoS ONE 6:e21049. https://doi.org/10.1371/journal.pone.0021049
Neifar M, Chouchane H, Najjari A et al (2019) Genome analysis provides insights into crude oil degradation and biosurfactant production by extremely halotolerant Halomonas desertis G11 isolated from Chott El-Djerid salt-lake in Tunisian desert. Genomics 111:1802–1814. https://doi.org/10.1016/j.ygeno.2018.12.003
Wells T, Ragauskas AJ (2012) Biotechnological opportunities with the β-ketoadipate pathway. Trends Biotechnol 30:627–637. https://doi.org/10.1016/j.tibtech.2012.09.008
Grund E, Denecke B, Eichenlaub R (1992) Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl Environ Microbiol 58:1874–1877. https://doi.org/10.1128/aem.58.6.1874-1877.1992
Acknowledgements
This study is based partly on the Ph.D. thesis of BM. Enuh. Besides, we thank Prof. Mutlu for providing the strain.
Funding
This work has been supported by Eskisehir Osmangazi University Scientific Research Projects Coordination Unit under grant number FDK-2022-2468.
Author information
Authors and Affiliations
Contributions
PAÇ and EBM developed and designed the study, EBM did the literature search and experiments and analysis, PAÇ revised the work critically. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflicts of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Enuh, B.M., Aytar Çelik, P. Genome Analysis of Halomonas elongata Strain 153B and Insights Into Polyhydroxyalkanoate Synthesis and Adaptive Mechanisms to High Saline Environments. Curr Microbiol 80, 18 (2023). https://doi.org/10.1007/s00284-022-03115-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03115-w