Abstract
Haloterrigena jeotgali is a halophilic archaeon within the family Natrialbaceae that was isolated from shrimp jeotgal, a traditional Korean salt-fermented food. A29T is the type strain of H. jeotgali, and is a Gram-negative staining, non-motile, rod-shaped archaeon that grows in 10 %–30 % (w/v) NaCl. We present the annotated H. jeotgali A29T genome sequence along with a summary of its features. The 4,131,621 bp genome with a GC content of 64.9 % comprises 4,215 protein-coding genes and 127 RNA genes. The sequence can provide useful information on genetic mechanisms that enable haloarchaea to endure a hypersaline environment.
Similar content being viewed by others
Introduction
An extremely halophilic archaeon, called a haloarchaeon, that is a member of the family Natrialbaceae [1] was isolated from various hypersaline environments such as soda and salt lakes, solar salterns, salt mines, salted soils, deep-sea brine, and various salt-fermented foods. Although high salinity is toxic to most cells, extreme halophiles are adapted to their hypersaline environments [2]. Most halophilic archaea require at least 1.5 M NaCl for growth and optimum growth occurs in the range of 3.1 to 3.4 M NaCl [3]. Since halophilic enzymes from the haloarchaea are generally considered to be active and stable at high salt concentrations, they have potential for biotechnological applications such as engineering for salt-resistant plants in agriculture, environmental bioremediation of organic pollutants and production of fermented foods. The genus Haloterrigena was first proposed by Ventosa et al. [4] with the reclassification of Halococcus turkmenicus as Haloterrigena turkmenica [4], and presently includes nine species: H. turkmenica [4], H. thermotolerans [5], H. longa , H. limicola [6], H. saccharevitans [7], H. hispanica [8], H. jeotgali [9], H. salina [10], and H. daqingensis [11], all of which are pleomorphic, Gram-negative staining, and red- or light pink-pigmented. However, the genus Haloterrigena is poorly characterized at the genome level.
A29T (= KCTC 4020T = DSM 18794T = JCM 14585T = CECT 7218T) is the type strain of H. jeotgali and was isolated from shrimp jeotgal, a traditional Korean salt-fermented food [9]. Although little is known about the roles of the haloarchaea during the fermentation process, the increasing genome information is expected to contribute to expansion of the understanding of their roles and halotolerant features. Here, we present a summary of the classification and features of H. jeotgali A29T along with the annotated genome sequence.
Organism information
Classification and features
A taxonomic analysis was conducted by comparing the H. jeotgali A29T 16S rRNA gene sequence with the most recent release of the EzTaxon-e database [12]. Phylogenetic relationships between strain A29T and closely related species were evaluated using MEGA6 program [13], and dendrograms were generated by the neighbor-joining [14], minimum evolution [15], and maximum likelihood [16] methods. A bootstrap analysis investigating the stability of the dendrogram was performed by obtaining a consensus tree based on 1,000 randomly generated trees. Strain A29T showed the highest level of the 16S rRNA gene similarity to H. thermotolerans PR5T (99.0 %), H. saccharevitans AB14T (98.3 %), H. limicola AX-7T (97.1 %), H. turkmenica 4kT (96.8 %), H. salina XH-65T (96.6 %), H. hispanica FP1T (96.1 %), H. longa ABH32T (94.9 %), and H. daqingensis JX313T (94.6 %). The DNA-DNA relatedness between strain A29T and the related strains H. thermotolerans PR5T, H. saccharevitans AB14T, and H. limicola AX-7T was 23.2 %, 22.0 %, and 17.9 %, respectively. The 16S rRNA gene sequence similarity data and DNA–DNA relatedness value of less than 70 % [17] suggested that strain A29T represents a distinct genospecies [9] (Table 1). The consensus phylogenetic tree based on the 16S rRNA gene sequences indicated that strain A29T was clustered in a branch with other species of the genus Haloterrigena (Fig. 1).
Phylogenetic tree based on the neighbor-joining (NJ) algorithm for the 16S rRNA gene sequences of strain A29T and closely related taxa. Numbers at the nodes indicate bootstrap values calculated using NJ/minimum evolution (ME)/maximum likelihood (ML) probabilities. Filled and open circles represent nodes recovered by both ME and ML methods or by either method, respectively. Methanospillum hangatei JF-1T served as an outgroup
H. jeotgali A29T is Gram-negative staining, non-motile, rod-shaped (0.4 μm wide and 1.0 μm long) (Fig. 2), and grows in irregular clusters. Colonies cultured on complex agar medium were light red, circular, and measured 0.5 mm in diameter after 7 days at 37 °C. Growth occurred in the presence of 10–30 % (w/v) NaCl at temperatures ranging from 17–50 °C and in the pH range of 6.5–8.5. Optimal conditions for growth were; a NaCl concentration of 15–20 % (w/v), a temperature ranging from 37–45 °C, and a pH of 7.0–7.5. The isolate was catalase-positive and oxidase-negative and did not reduce nitrate to nitrite. Mg2+ was not required for growth. Cell lysis occurred in distilled water. This strain was able to hydrolyze casein and Tween 80 but not starch, gelatin, urea, or DNA. Anaerobic growth occurred in the presence of nitrate but not of sulfate, thiosulfate, dimethyl sulfoxide, or trimethylamine N-oxide. Fructose, lactose, and acetate—but not sucrose, glucose, citrate, or formate—were utilized as carbon and energy sources. Acid was not produced from fructose, lactose, acetate, sucrose, glucose, citrate, or formate. Strain A29T was resistant to bacitracin, penicillin, ampicillin, chloramphenicol, and erythromycin, but was sensitive to novobiocin, anisomycin, and aphidicolin. The major polar lipids were phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, and mannose-2,6-disulfate(1–2)-glucose glycerol diether [9].
Genome sequencing and annotation
Genome project history
H. jeotgali strain A29T genome was sequenced to obtain information regarding mechanism(s) or molecule(s) that confer adaption to a hypersaline environment and to identify the primary structure of potentially novel halophilic enzymes with relatively low similarity to those in the sequence database. The genome project and sequence were deposited in the Genomes OnLine Database [18] and GenBank (JDTG00000000), respectively. Sequencing and annotation were performed by ChunLab Inc. (Seoul, Korea). Project information and associated MIGS version 2.0 compliance levels [19] are shown in Table 2.
Growth conditions and genomic DNA preparation
H. jeotgali A29T was grown aerobically in DSM Medium 954 at 37°C. Genomic DNA was extracted and purified using a G-spin™ DNA extraction kit (iNtRON Biotechnology, Sungnam, Korea) according to the manufacturer’s instructions.
Genome sequencing and assembly
The genome of H. jeotgali A29T was sequenced from a total of 9,473,809 quality-trimmed sequencing reads (700.5-fold coverage) that combined 6,797,702 reads (473.8-fold coverage) from the Illumina MiSeq. 300 bp paired-end library (Illumina, San Diego, CA, USA); 2,617,102 reads (181.1-fold coverage) obtained using an Ion Torrent Personal Genome Machine (PGM) 318v2 chip (Life Technologies, Carlsbad, CA, USA); and 59,005 reads (45.7-fold coverage) from a PacBio RS 10 kb library (Pacific Biosciences, Menlo Park, CA, USA). Illumina and PGM data were assembled de novo with CLC Genomics Workbench 6.5.1 (CLC bio, Boston, MA, USA) and PacBio data were assembled with the HGAP2 algorithm in SMRT Analysis 2.1 (Pacific Biosciences). Resultant contigs were assembled with CodonCode Aligner 3.7 (CodonCode Corporation, Centerville, MA, USA). The final assembly yielded three scaffolds with 20 contigs spanning 4.1 Mb.
Genome annotation
Open reading frames of the assembled genome were predicted using the Integrated Microbial Genomes-Expert Review platform as part of the Joint Genome Institute genome annotation pipeline [20]. Additional gene prediction and functional annotation were achieved using the Rapid Annotation using Subsystem Technology pipeline. Predicted ORFs were compared during gene annotation using NCBI Clusters of Orthologous Groups [21], Pfam [22], and EzTaxon-e [12] databases. rRNA and tRNA genes were identified using RNAmmer 1.2 [23] and tRNAscan-SE 1.23 [24] tools, respectively. Genomic features were visualized with CLgenomics 1.06 (ChunLab Inc.).
Genome properties
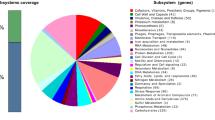
The draft genome sequence of H. jeotgali A29T was 4,131,621 bp and comprised three scaffolds including 20 contigs, and had a GC content of 64.9 % (Fig. 3 and Table 3). Of the 4,342 predicted genes, 4,215 were protein-coding and 2,636 ORFs (60.7 %) were assigned putative functions, whereas the remaining genes were annotated as hypothetical proteins. The genome contained 127 ORFs assigned to RNA genes, including 47 predicted for tRNA, 14 for rRNA (five 5S, two 16S, and seven 23S), and 66 for miscellaneous RNA (one archaeal signal recognition particle; five for the HgcC family; one archaeal RNA P; and 59 clustered regularly interspaced short palindromic direct repeat elements). The distribution of genes across COG functional categories is presented in Table 4.
Graphical circular map of the H. jeotgali A29T genome. RNA genes (red, tRNA and blue, rRNA) and genes on the reverse and forward strands (colored according to COG categories) are shown from the outside to the center. The inner circle shows the GC skew; yellow and blue indicate positive and negative values, respectively. GC content is indicated in red and green
Conclusions
H. jeotgali A29T encoded the genes associated with the mechanisms of salinity tolerance, biosynthesis and transport of compatible solutes such as glycine betaine (N,N,N-trimethylglycine) (choline sulfatase, choline dehydrogenase, betaine reductase, and glycine betaine transporter OpuD), ion exclusion using cation (Mg2+ and Cu2+) transport and K+ transport and Na+/H+ antiporter systems. The sequences may contribute to expansion of our knowledge of complex osmoregulation mechanism of the haloarchaea that should facilitate biotechnological applications of the haloarchaea and provide useful information on genetic mechanisms that enable haloarchaea to endure hypersaline environments.
Abbreviations
- ME:
-
Minimum evolution
- ML:
-
Maximum likelihood
- NJ:
-
Neighbor-joining
- PGM:
-
Personal Genome Machine
References
Gupta RS, Naushad S, Baker S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov. Int J Syst Evol Microbiol. 2015;65:1050–69.
Grant WD. Life at low water activity. Philos Trans R Soc Lond B Biol Sci. 2004;59:1249–66.
Oren A. Sensitivity of selected members of the family Halobacteriaceae to quinolone antimicrobial compounds. Arch Microbiol. 1996;165:354–8.
Ventosa A, Gutierrez MC, Kamekura M, Dyall-Smith ML. Proposal to transfer Halococcus turkmenicus, Halobacterium trapanicum JCM 9743 and strain GSL-11 to Haloterrigena turkmenica gen. nov., comb. nov. Int J Syst Bacteriol. 1999;49:131–6.
Montalvo-Rodriguez R, Lopez-Garriga J, Vreeland RH, Oren A, Ventosa A, Kamekura M. Haloterrigena thermotolerans sp. nov., a halophilic archaeon from Puerto Rico. Int J Syst Evol Microbiol. 2000;50:1065–71.
Cui HL, Tohty D, Zhou PJ, Liu SJ. Haloterrigena longa sp. nov. and Haloterrigena limicola sp. nov., extremely halophilic archaea isolated from a salt lake. Int J Syst Evol Microbiol. 2006;56:1837–40.
Xu XW, Liu SJ, Tohty D, Oren A, Wu M, Zhou PJ. Haloterrigena saccharevitans sp. nov., an extremely halophilic archaeon from Xin-Jiang, China. Int J Syst Evol Microbiol. 2005;55:2539–42.
Romano I, Poli A, Finore I, Huertas FJ, Gambacorta A, Pelliccione S, et al. Haloterrigena hispanica sp. nov., an extremely halophilic archaeon from Fuente de Piedra, southern Spain. Int J Syst Evol Microbiol. 2007;57:1499–503.
Roh SW, Nam YD, Chang HW, Kim KH, Sung Y, Kim MS, et al. Haloterrigena jeotgali sp. nov., an extremely halophilic archaeon from salt-fermented food. Int J Syst Evol Microbiol. 2009;59:2359–63.
Gutierrez MC, Castillo AM, Kamekura M, Ventosa A. Haloterrigena salina sp. nov., an extremely halophilic archaeon isolated from a salt lake. Int J Syst Evol Microbiol. 2008;58:2880–4.
Wang S, Yang Q, Liu ZH, Sun L, Wei D, Zhang JZ, et al. Haloterrigena daqingensis sp. nov., an extremely haloalkaliphilic archaeon isolated from a saline-alkaline soil. Int J Syst Evol Microbiol. 2010;60:2267–71.
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–21.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol. 1992;9:945–67.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–4.
Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, et al. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:346–54.
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7.
Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–8.
Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:222–30.
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64.
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9.
Garrity GM, Holt JG. Phylum AII. Euryarchaeota phy. nov. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology, vol. 2. 2nd ed. New York: Springer; 2001. p. 211.
Editor L. Validation List no. 85. Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2002;52:685–90.
Grant WD, Kamekura M, McGenity TJ, Ventosa A. Class III. Halobacteria class nov. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology, vol. 2. 2nd ed. New York: Springer; 2001. p. 294–334.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2012R1A1A2040922 and 2014R1A1A1002980) and a Korea Basic Science Institute NAP grant (T34780).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KJY and HSS carried out the microbial cultivation and DNA isolation. ITC, MHL, SWR, BYK, YJC and DWK participated in the sequence analyses. ITC, MHL and SWR drafted the manuscript. M-JS, J-KR and CY helped to draft the manuscript. SWR and Y-DN conceived of the study and participated in its design. H-JC and J-SC participated in its design and coordination. All authors read and approved the final manuscript.
In-Tae Cha and Mi-Hwa Lee contributed equally to this work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cha, IT., Lee, MH., Kim, BY. et al. Genome sequence of the haloarchaeon Haloterrigena jeotgali type strain A29T isolated from salt-fermented food. Stand in Genomic Sci 10, 49 (2015). https://doi.org/10.1186/s40793-015-0047-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40793-015-0047-4